Many genome sequences in public databases are used in metagenomic studies to understand the human microbiome. Unfortunately, many of the taxonomic identities of these genomes are either incorrect or incomplete. Since bacterial species are now defined by comparing genome sequences, having the genome sequence should lead us to the correct identification of any organism at the species, or even subspecies level. The only prerequisite condition is the availability of relevant type strain genomes. In this series of blogs, we will present the correct taxonomic identification of some misidentified genomes frequently used in the publications. These genome sequences were identified using our TrueBac IDTM service.
The case of Ruminococcus sp. 5_1_39BFAA
This strain likely belongs to the species that are widely found in the human gut microbiome as it has appeared in many publications.
Select Publications reporting Ruminococcus sp. 5_1_39BFAA (a few of many)
- Li et al. (2016) Science. Durable coexistence of donor and recipient strains after fecal microbiota transplantation
- Guo et al. (2017) Cell. Discovery of reactive microbiota-derived metabolites that inhibit host proteases
- Feng et al. (2015) Nature Communications. Gut microbiome development along the colorectal adenoma–carcinoma sequence
- Dubinkina (2017) Microbiome. Links of gut microbiota composition with alcohol dependence syndrome and alcoholic liver disease
Taxonomic information in the public database
In NCBI and other databases, it is labeled as “Ruminococcus sp.” under the genus Ruminococcus and the family Ruminococcaceae.
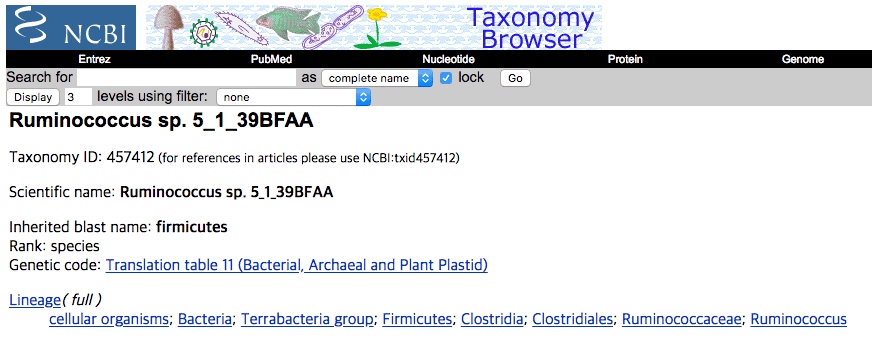
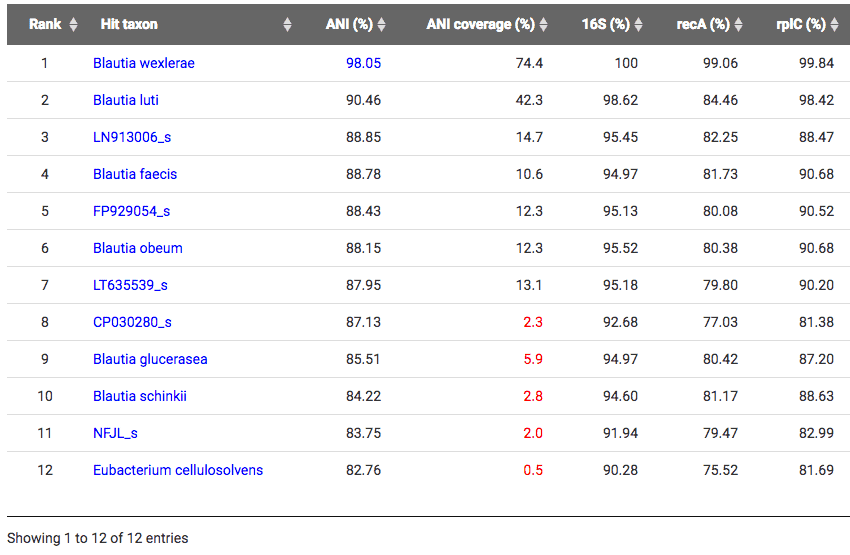
Correct taxonomic identification of Ruminococcus sp. 5_1_39BFAA
With the TrueBac ID’s algorithm, this genome was identified as a strain of Blautia wexlerae in the genus Blautia and the family Lachnospiraceae by average nucleotide identity (ANI) value of 98.05% to the type strain of Blautia wexlerae. ANI values of 95~96% are considered as the cut-off for the species boundary [Learn more]. Clearly, it is not a member of Ruminococcus. To browse this genome at EzBioCloud database, visit here. Below is a screen-shot of the TrueBac ID result showing what is now classified as Blautia wexlerae 5_1_39BFAA.
Follow the EzBioCloud’s blog using Twitter @ezbiocloud
Sept. 17, 2018