This data field contains the original strain labels from genome projects obtained from public databases (e.g. Genbank) or one that was entered by a user in the case of private genome projects. CJ Bioscience uses the combination of fast search algorithms and robust OrthoANI calculation to identify each public or private genomes. Often, the taxonomic identity of a genome is different from what’s labelled by public databases or creator of the data. In EzBioCloud, we uses correct taxonomic names, wherever possible. However, the original labels are also preserved and provided so users are not confused.
Below is an example of wrong labels in Genbank:
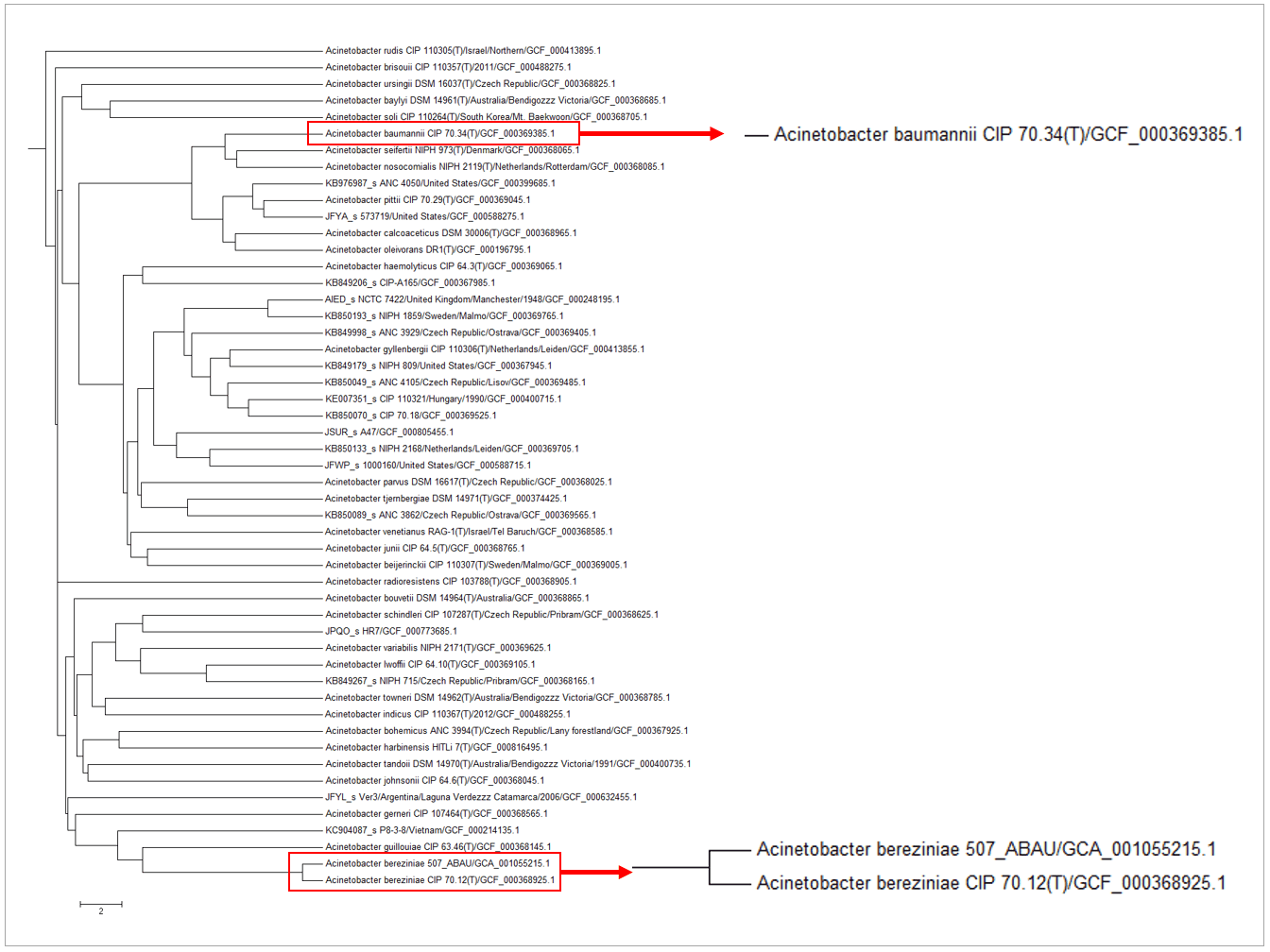
Genome with the accession of GCF_001055215.1 is labeled as “Acinetobacter baumannii” in Genbank database. After phylogenomic comparison with type/reference strains using OrthoANI (see the below figure), it is clear that this genome should be named “Acinetobacter bereziniae“. The OrthoANI value between the type strain of Acinetobacter bereziniae and GCF_001055215.1 is 98.05%, which is higher than the species boundary proposed by Lee et al. (2015).
Last updated on May 13th, 2016 (EK)


