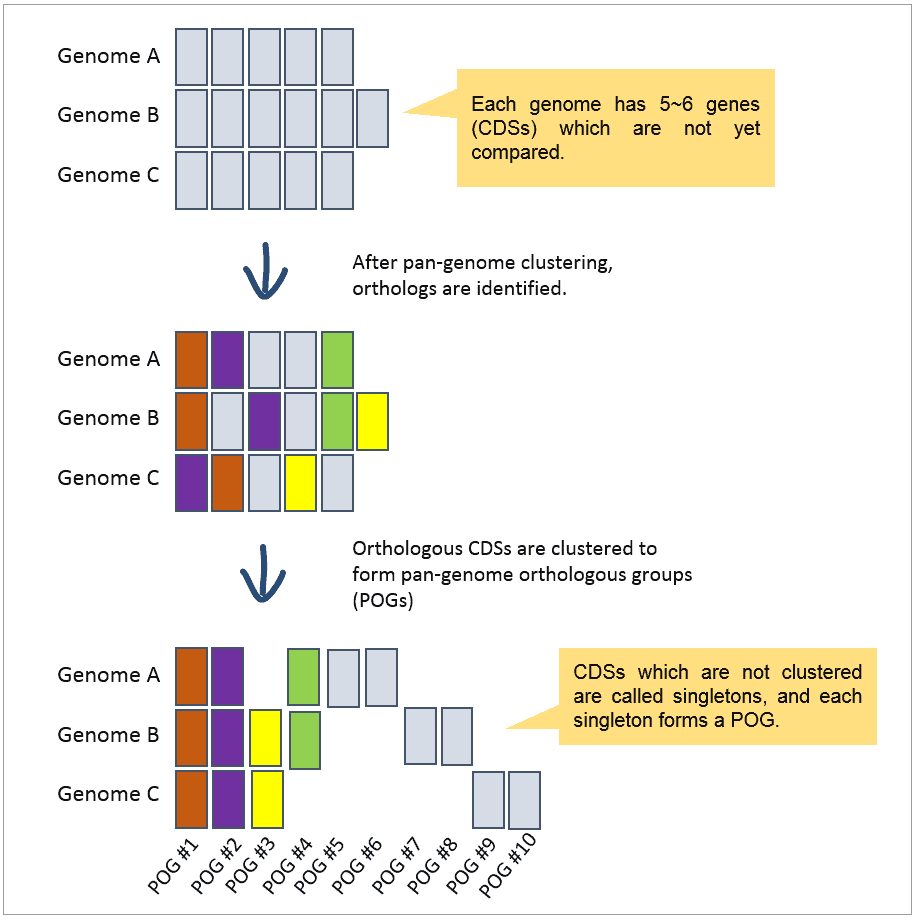
Pan-genome orthologous group (POG) refers to “a group of orthologous genes (orthologs)” that are identified from multiple genomes. The below figure illustrates how POGs are identified.
In this example, a total of 10 POGs were identified from clustering 16 genes (CDSs) in 3 genomes.
- Two POGs (#1 & 2) comprise the core-genome as they are present in all genomes.
- Six POGs (#5~10) contain single CDS, called “singleton”.
- Pan-genome of these 3 genomes consists of 10 POGs.
POGs are the basic unit of core- and pan-genome.
Last updated on May 10th, 2016 (EK)