This tutorial will explain how to process a 16S sequence into a phylogenetic tree with EzBioCloud and EzEditor2.
1. Open www.ezbiocloud.net and go to the “Identify” page. Login is necessary so you may need to register which is free.
2. We will use a 16S rRNA gene sequence obtained from a human fecal sample. It was amplified by universal bacterial PCR primers 27F and 1492R, and sequenced by the Pacific Biosciences (PacBio) RS1. Full-length, accurate 16S sequences can be obtained using the circular consensus sequence (ccs) method of PacBio’s single molecule sequencing.
Copy the below FASTA format to the clipboard of your computer.
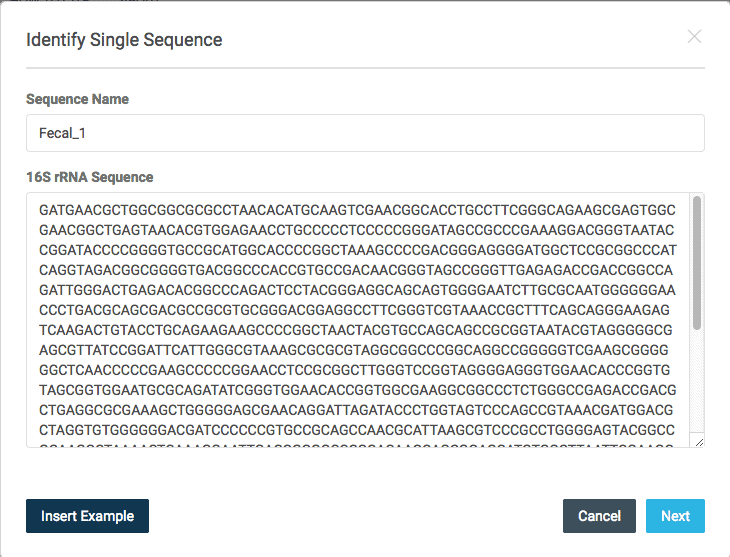
>Fecal_1 GATGAACGCTGGCGGCGCGCCTAACACATGCAAGTCGAACGGCACCTGCCTTCGGGCAGAAGCGAGTGGCGAACGGCTGAGTAACACGTGGAGAACCTGCCCCCTCCCCCGGGATAGCCGCCCGAAAGGACGGGTAATACCGGATACCCCGGGGTGCCGCATGGCACCCCGGCTAAAGCCCCGACGGGAGGGGATGGCTCCGCGGCCCATCAGGTAGACGGCGGGGTGACGGCCCACCGTGCCGACAACGGGTAGCCGGGTTGAGAGACCGACCGGCCAGATTGGGACTGAGACACGGCCCAGACTCCTACGGGAGGCAGCAGTGGGGAATCTTGCGCAATGGGGGGAACCCTGACGCAGCGACGCCGCGTGCGGGACGGAGGCCTTCGGGTCGTAAACCGCTTTCAGCAGGGAAGAGTCAAGACTGTACCTGCAGAAGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCGAGCGTTATCCGGATTCATTGGGCGTAAAGCGCGCGTAGGCGGCCCGGCAGGCCGGGGGTCGAAGCGGGGGGCTCAACCCCCGAAGCCCCCGGAACCTCCGCGGCTTGGGTCCGGTAGGGGAGGGTGGAACACCCGGTGTAGCGGTGGAATGCGCAGATATCGGGTGGAACACCGGTGGCGAAGGCGGCCCTCTGGGCCGAGACCGACGCTGAGGCGCGAAAGCTGGGGGAGCGAACAGGATTAGATACCCTGGTAGTCCCAGCCGTAAACGATGGACGCTAGGTGTGGGGGGACGATCCCCCCGTGCCGCAGCCAACGCATTAAGCGTCCCGCCTGGGGAGTACGGCCGCAAGGCTAAAACTCAAAGGAATTGACGGGGGCCCGCACAAGCAGCGGAGCATGTGGCTTAATTCGAAGCAACGCGAAGAACCTTACCAGGGCTTGACATATGGGTGAAGCGGGGGAGACCCCGTGGCCGAGAGGAGCCCATACAGGTGGTGCATGGCTGTCGTCAGCTCGTGTCGTGAGATGTTGGGTTAAGTCCCGCAACGAGCGCAACCCCCGCCGCGTGTTGCCATCGGGTGATGCCGGGAACCCACGCGGGACCGCCGCCGTCAAGGCGGAGGAGGGCGGGGACGACGTCAAGTCATCATGCCCCTTATGCCCTGGGCTGCACACGTGCTACAATGGCCGGTACAGAGGGATGCCACCCCGCGAGGGGGAGCGGATCCCGGAAAGCCGGCCCCAGTTCGGATTGGGGGCTGCAACCCGCCCCATGAAGTCGGAGTTGCTAGTAATCGCGGATCAGCATGCCGCGGTGAATGCGTTCCCGGGCCTTGTACACACCGCCCGTCACACCACCCGAGTCGTCTGCACCCGAAGTCGCCGGCCCAACCGAGAGGGGGGAGGCGCCGAAGGTGTGGAGGGTGAGGGGGGTG
3. At ezbiocloud Identify page, Select [Identify single sequence] and paste the sequence into the area shown below.
Submit your identification request and wait for a minute or so.
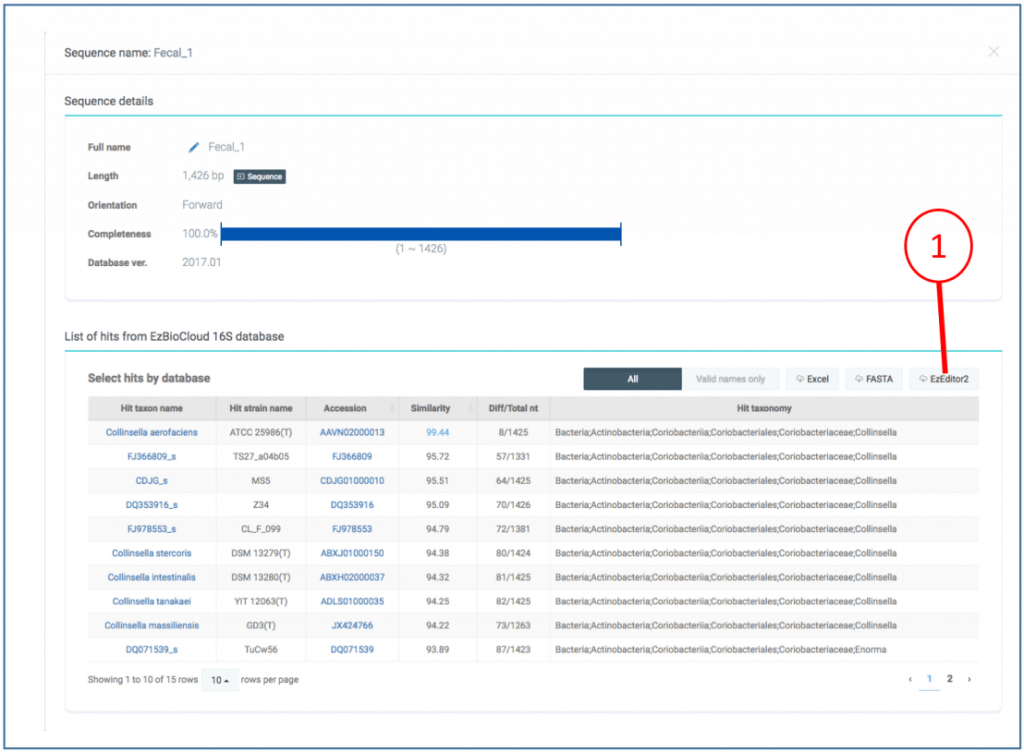
The EzBioCloud server will finish your job request as:
(1) Our query sequence showed 99.44% similarity to Collinsella aerofaciens.
(2) Explore more details by clicking an icon indicated by (2)
(1) Download the data file for EzEditor2 program. This file contains all the hit and query sequences. Hit sequences are already aligned manually by CJ Bioscience’s staff. Now, we will use EzEditor2 program to align our query sequence against hit sequences with secondary structure information.
Your query sequence is placed at the bottom of the alignment. The most closely related hit sequence is placed right above your query sequence.
Please use the following tutorials for manual alignment using secondary structure information and phylogenetic analysis.
Go to “Working with 16S rRNA sequences“
Go to “Phylogenetic analysis using EzEditor2”
Last modified on February 1, 2017 (JC)