The genome sequence that is labeled as “Lachnospiraceae bacterium 5_1_63FAA” has been included in most microbiome taxonomic profiling software tools, and has therefore appeared in many publications. The following list is a small sampling:
- Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. 2016, Cell 167:1125–1136
- Role of gut microbiota, bile acids and their cross‐talk in the effects of bariatric surgery on obesity and type 2 diabetes. 2018. J Diabetes Investig. 2018 Jan;9(1):13-20.
- Changes of the human gut microbiome induced by a fermented milk product. 2014. Scientific Reports 4: 6328
- Total Lipopolysaccharide from the Human Gut Microbiome Silences Toll-Like Receptor Signaling. 2017. mSystems 2(6):e00046-17.
- A Phylogenomic View of Ecological Specialization in the Lachnospiraceae, a Family of Digestive Tract-Associated Bacteria. 2014. Genome Biol Evol. 2014 Mar; 6(3): 703–713.
Since this genome is cited in many studies on the human gut microbiome, this species is likely to be a major component of us.
This genome was sequenced in 2011 and assigned as an unclassified species in the family Lachnospiraceae.
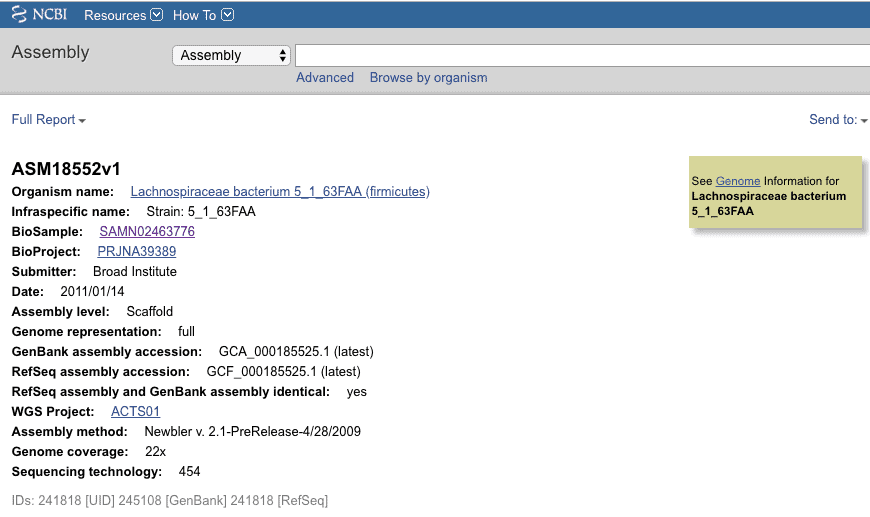
I used our TrueBac ID system with it’s up-to-date, taxonomically curated database to identify this genome sequence. The results show that this genome belongs to Anaerostipes hadrus. This species was first published as ‘Eubacterium hadrum’ in 1976, then reclassified in the genus Anaerostipes in 2013 (Allen-Vercoe et al. 2013).
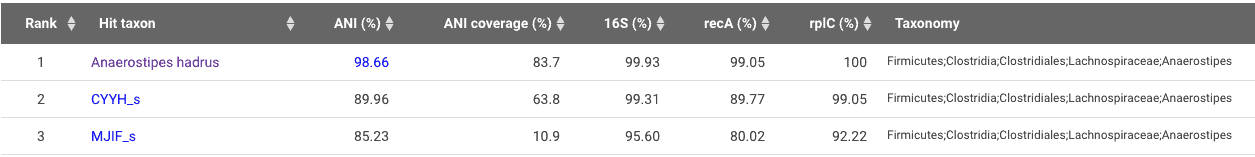
The ANI value is sufficiently higher than the proposed cutoff (95~96%) by Chun et al. (2018). Please also note that there are two other genomospecies closely related to 5_1_63FAA in this genus, namely, CYYH_s and MJIF_s.
I hope to see more of Anaerostipes hadrus 5_1_63FAA in the publications instead of Lachnospiraceae bacterium 5_1_63FAA in future.
by Jon Jongsik Chun
October 28, 2018