A pan-genome is a union of the protein-coding genes (=CDS) in a given set of genomes. Generally, building a pan-genome starts from a set of annotated genomes (see below).
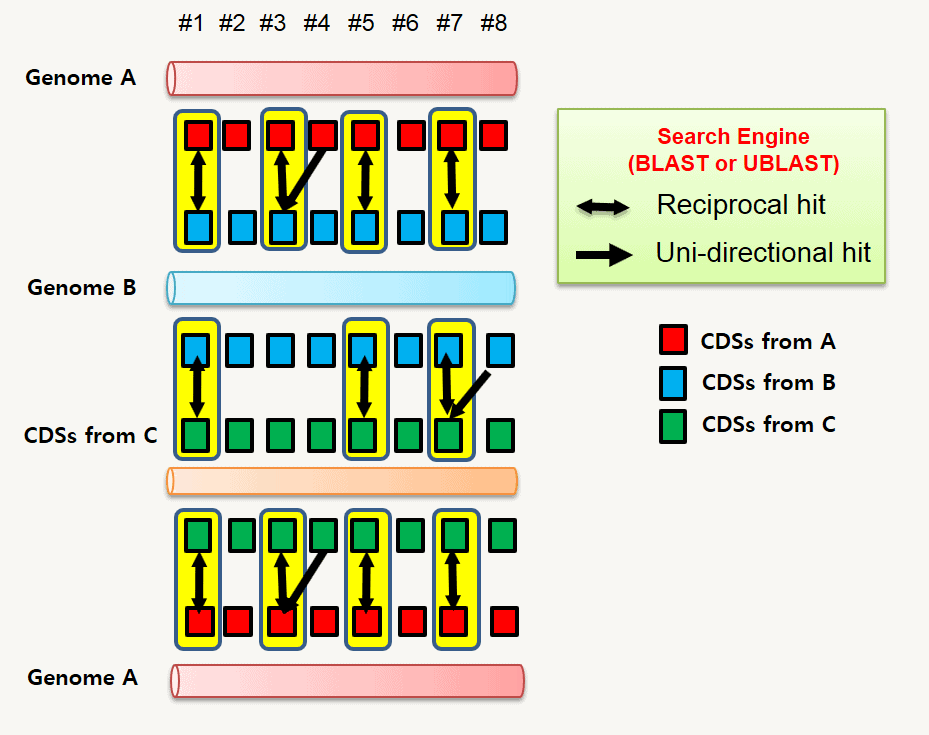
Then, each genome pair is subjected to a homology search using BLAST or uBLAST programs. When CDS #1 in genome A matches best to CDS #33 of genome B and vice versa, two CDSs are said to be “reciprocally best hit (RBH)” (orthologs). This process is carried out for all possible genome pairs. If we analyze 10 genomes, it would be 100 (10×10) comparisons minus self comparison. So, it would be 90 pair-wise comparisons for the data set containing 10 genomes. See here for more details about how to detect orthologs.
The below figure shows the results of pairwise reciprocal searches among 3 genomes (A,B,C).
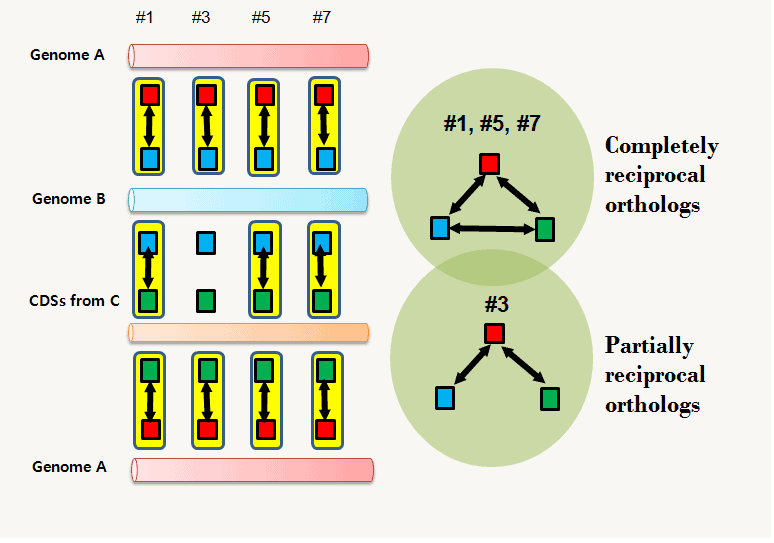
From the above example, we can identify 4 clusters of orthologous CDSs (=orthologs). If we just take the “reciprocally best hits” (see below),
CDS #1, 5 and, 7 show “complete” reciprocity, whereas CDS #3 is only “incompletely or partially” reciprocal among the 3 genomes. In the latter case, each CDS from the three genomes can still be connected (CDS #3 from genome A in red connects CDSs from B and C genomes even though they are not directly connected) .
In EzCgDb, pan-genome is a collection of orthologous groups that include partially reciprocal orthologs (as in the above figure), and each group is called “pan-genome orthologous group (POG)“. If there are N genomes in a data set, some POGs are found in all N genomes (comprising core-genome with 100% cutoff) whereas some POGs are found only in a single genome (=singletons).
[Update info ver1.1]
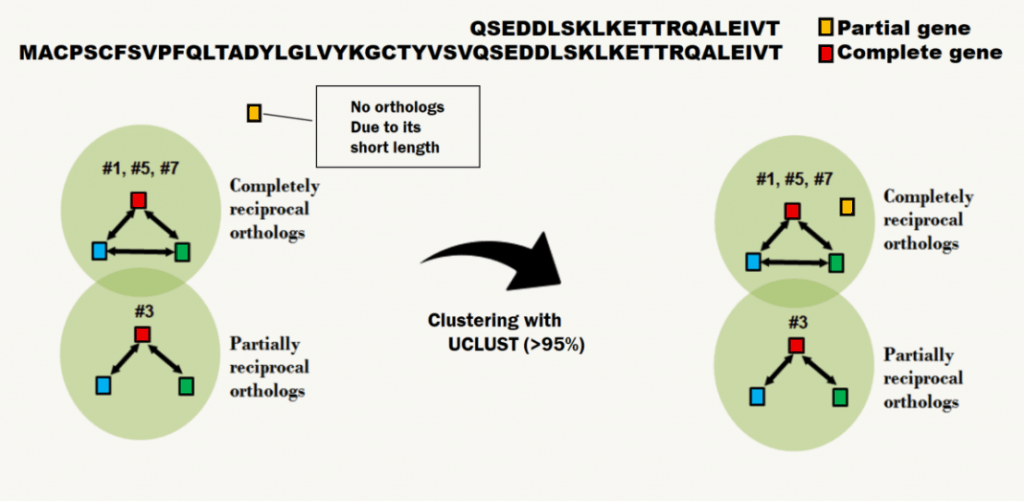
The downfall of getting a pan-genome happens when there is a partial gene present in the genome. When there is a partial gene, the reciprocity will not be formed due to its short length. As a result, the gene remains as a singleton and considered as a different gene, even though it has 100% sequence identity to the orthologous group. To avoid such issue, UCLUST is performed to bind a partial gene to a proper orthologous group, which basically regroups the genes if it has 95% identity (see below). The details about UCLUST algorithm is provided in here.
Last updated on Aug 8th, 2016. (SH)