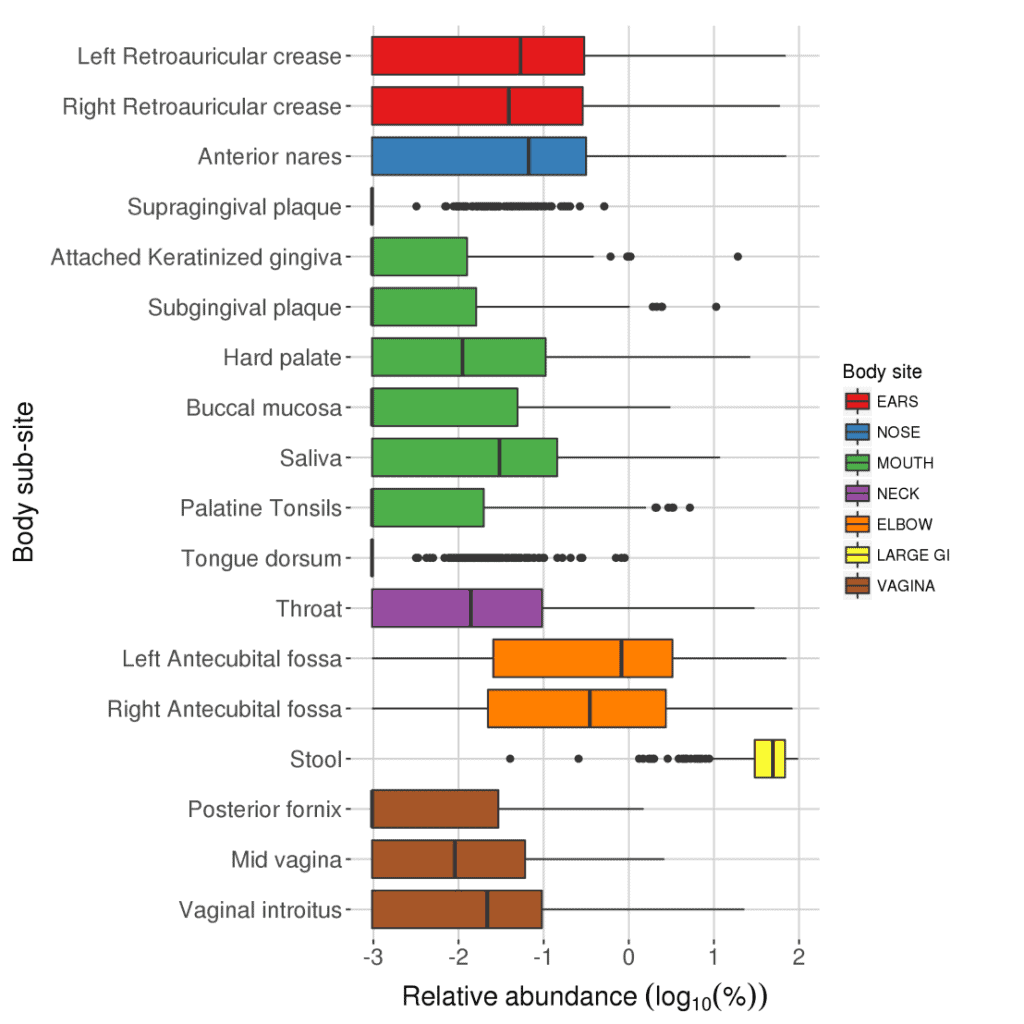
Taxonomic profile of human microbiome at EzBioCloud.net
EzBioCloud 16S database provides a comprehensive taxonomic backbone of Bacteria. Here we applied the sophisticated bioinformatics pipeline for a large dataset of the U.S. human microbiome project, which allows us to recognize major bacterial inhabitants of our body.
- Data source: U.S. NIH human microbiome project (http://hmpdacc.org/)
- NGS platform: Roche 454
- Sequencing regions of 16S gene: various
- Human subjects: 242 healthy adults sampled at 15 or 18 body sites
- Bioinformatics pipeline used: CJ Bioscience’s microbiome taxonomic profiling (MTP) pipeline described more details at here.
- The number of samples used in the calculation:
| Body sites | Samples | |
| 1 | Anterior nares | 433 |
| 2 | Attached Keratinized gingiva | 545 |
| 3 | Buccal mucosa | 536 |
| 4 | Hard palate | 502 |
| 5 | Left Antecubital fossa | 280 |
| 6 | Left Retroauricular crease | 458 |
| 7 | Mid vagina | 216 |
| 8 | Palatine Tonsils | 563 |
| 9 | Posterior fornix | 216 |
| 10 | Right Antecubital fossa | 284 |
| 11 | Right Retroauricular crease | 487 |
| 12 | Saliva | 487 |
| 13 | Stool | 574 |
| 14 | Subgingival plaque | 558 |
| 15 | Supragingival plaque | 575 |
| 16 | Throat | 520 |
| 17 | Tongue dorsum | 570 |
| 18 | Vaginal introitus | 244 |
| Total | 8048 |
Statistics used:
(Example of the genus Bacteroides)
| Mean occurrence (%): | Mean occurrence of the taxon, calculated from all samples |
|---|---|
| Max occurrence (%): | Maximum occurrence found among all samples |
| Samples found (%): | Portion of samples in which the taxon was found |
Last modified Jan. 4, 2016 (JC)