There is significant confusion between bacterial type strains and reference strains. The type strain is defined in the context of formal bacterial taxonomy whereas reference strain means that the strain is widely used within the community or adopted by a formally recognized institution such as FDA and CDC. Below is a summary of these two terms:
[wp_table id=8846/]
Scientifically, when an isolate is identified as the species Escherichia coli, it should be based on the comparison against the type strain (Escherichia coli ATCC 11775T) but not any other strain including the famous Escherichia coli K12 MG1665.
How to obtain the type strains
The type strains of the species with valid names should be available to anyone for comparative analysis (for a reasonable fee). These are maintained as live cultures and can be obtained from various culture collections. A list of culture collections can be found at website of the World Federation of Culture Collections. If you are looking for the type strain of a particular species, the StrainInfo database is a useful source. For example, the information about the type strain of Escherichia coli can be obtained here. In theory, the type strain of any species with a valid name should be available for study. I said “in theory” because there are problems and exceptions in some cases.
How to obtain the genome sequence of a type strain
In modern bacterial taxonomy, classification and identification are mainly based on the pairwise comparison of genome sequences between the type strain and a strain under consideration. It is therefore of premium importance that the genome sequences of type strains are available for comparison. As for other nucleotide sequences, these data are deposited in the primary DNA sequence databases, i.e., NCBI, EMBL-EBI, and DDJB. There are now many genome sequences of the type strains, but one should be cautious before using these for any study. One must remember that the responsibility for authentication and validation of genome sequence data in these primary public databases, falls on the original depositors to the database, and not on the database curators. There are many cases where wrong data is labeled as those of the type strains. If these are used for classification and identification, then the results are fundamentally incorrect.
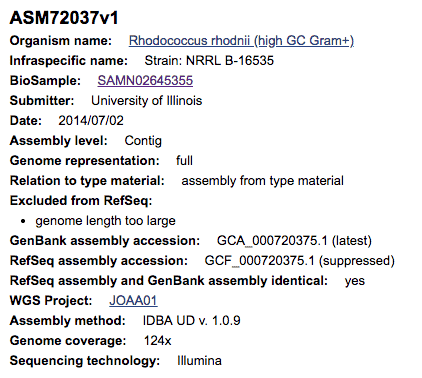
For example, the genome sequence of Rhodococcus rhodnii NRRL B-16535 with the NCBI accession GCF_000720375.1 (see the screenshot above), is labeled as that of the type strain. The strain designation NRRL B-16535 indicates that it is a type strain [Learn more]. After careful taxonomic investigation and curation, it becomes clear that this genome is not of the type strain of Rhodococcus rhodnii, but rather belongs to the genus Kibdelosporangium [Learn more]. Due to the lack of genome data for the type strains of Kibdelosporangium spp., we can identify this genome only up to the genus level (at the time of this writing).
If you have quality-controlled genome sequences of correct type strains of all known species, the identification of any bacterial isolate should be straightforward, in theory, with 100% accuracy.
Usages of “type strain” and “reference strain” in EzBioCloud and TrueBac TM databases
In the EzBioCloud public database and TrueBac TM database, both of the terms, type strain and reference strain, are used.
- Type strain: This is literally the type strains that have a standing in the formal nomenclature.
- Reference strain: This term is used for several cases.
- Species without proposed type strains: Some species, before the time of the publication of Approved Lists of Bacterial Names, do not have a designated type strain available from Culture Collections. For example, Wolbachia pipientis is a bacterial species that is an endosymbiont of the host mosquito, Culex pipiens. The type strain of this species could not be designated and deposited to culture collections as it cannot be grown as a pure culture. If this species is described in the present day, it will not be proposed as a species with a valid name, but rather, as a form of Candidatus. Nonetheless, we know that this species does exist in nature and sequences of its 16S rRNA gene and genome are available. In this case, the “reference” sequences are designated and used in our databases.
- Species without available type strains: For some species, the type strains are not available in any culture collections as they are lost over time. We used “reference” strains if there are available representative sequences for those species.
- Candidatus: Candidatus concept is not a part of the formal nomenclature, but was proposed for the cases where pure culture cannot be obtained (e.g. obligate symbionts). Most of Candidatus species are proposed without designation of the type strain. In this case, we assign 16S or/and genome sequences as “reference” instead of “type”.
- Genomospecies: This term represents the tentatively new species that were never officially described (as effective publications) [Learn more]. As more genome sequences are deposited to the primary sequence databases, more genomospecies are found. In our databases, genomospecies are identified using genome sequences and given a unique name. This helps to organize and understand the species diversity of Bacteria. For example, NCBI accession GCA_000369525.1 represents a new species in the genus Acinetobacter, so we named it the genomospecies “KB850070_s” for our database. After this, we were able to find three more genome sequences that belong to the genomospecies KB850070_s [Learn more]. Someday, someone may describe this new species in a formal way. Until then, the use of the genomospecies can improve the efficiency, quality and accuracy of ecology, metagenomics, and microbiome studies. A genomospecies is designated using the genome sequence.
The EzBioCloud team / Last edited on April 25, 2018