CJ Bioscience/EzBiome has a new platform that we developed to cover most of the analysis needed for CoVID. EzCOVID19 focuses on the rapid detection, identification, and characterization of the SARS-CoV-2 virus. We do this by:
- Allowing you to use multiple sample starting types (i.e. raw metagenomic, metatranscriptomic, RNA-seq, or isolates).
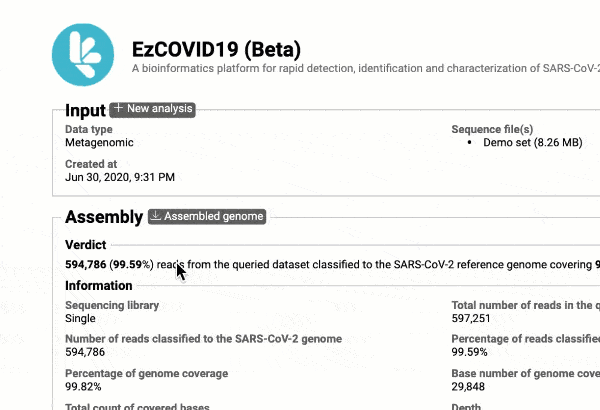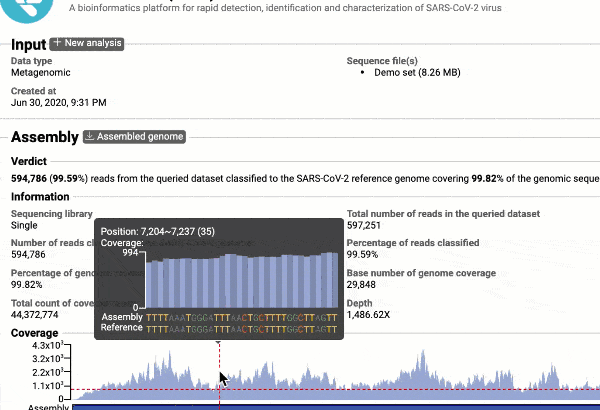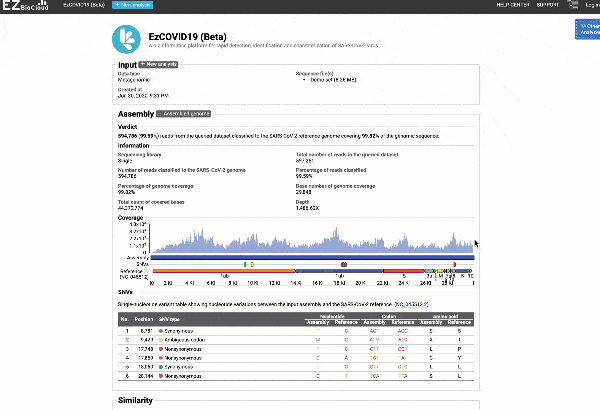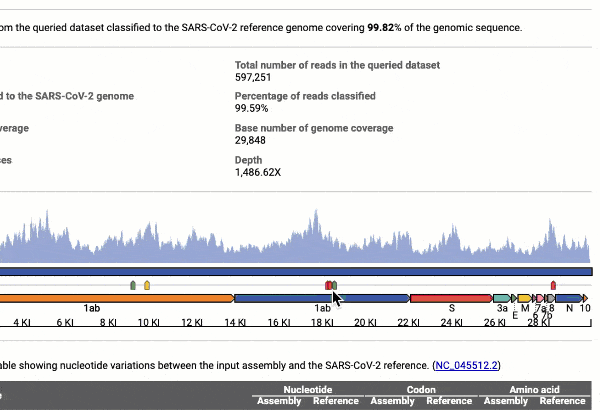
- Providing you with a consensus genome assembly along with statistics related to genome coverage, depth metrics, and coverage plots with single nucleotide variant (SNV) relative to the reference SARS-CoV-2 genome.
- Detection of most similar genomes available in the reference databases based on alignment statistics and SNVs, including a maximum likelihood or parsimony-based similarity tree decorated with SNV profiles.
- Classification or typing of the queried genome using EzBioCloud’s SNP based classification scheme of SARS-CoV-2 variants, including an evolutionary analysis of the detected SARS-CoV-2 type along with other types observed among publicly available SARS-CoV-2 genomes
Get the complete coverage information of the entire viral genome
In addition to the relationship to concurrently circulated publicly available SARS-CoV-2 genomes, EzCOVID19 enables characterization and typing of the entire viral genome, when the user obtains adequate coverage of the genome.
Graph and table with detected variants
See a detailed graphical breakdown of the variants as well as a table format highlighting the calls.
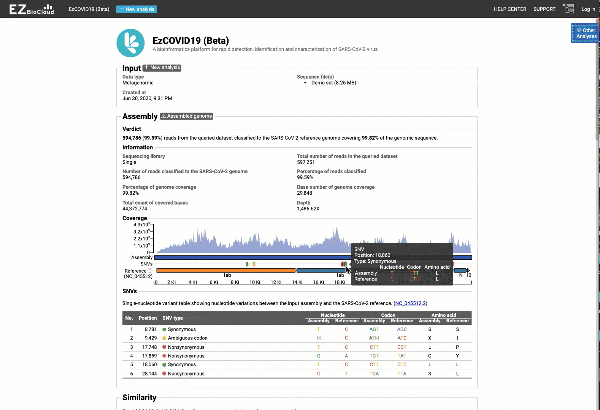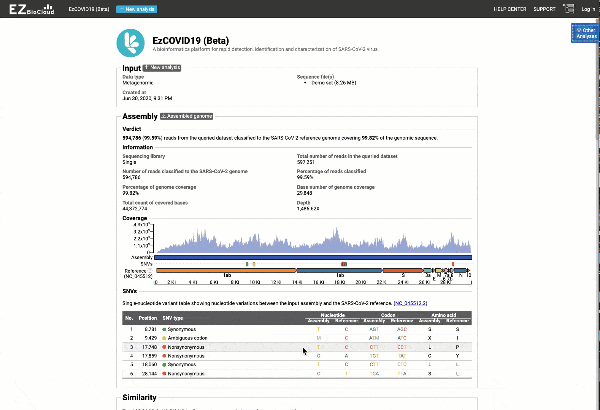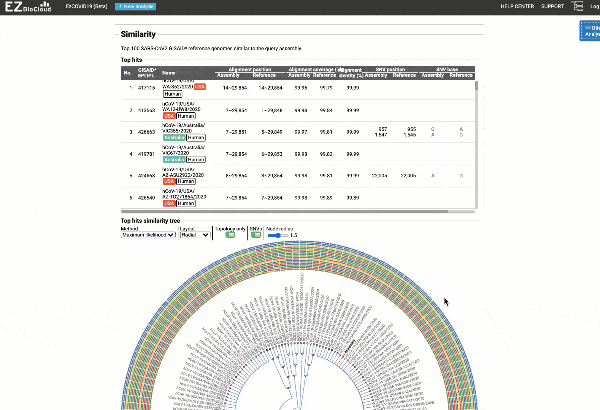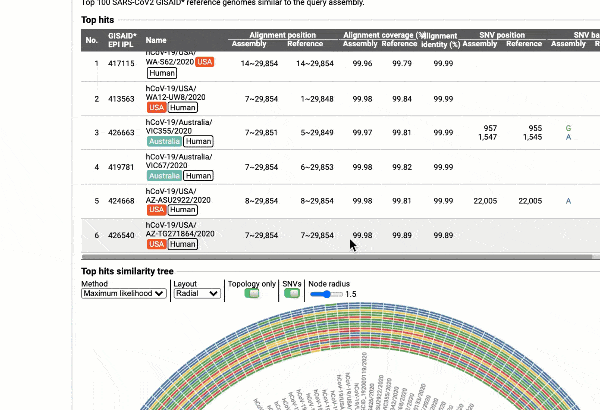
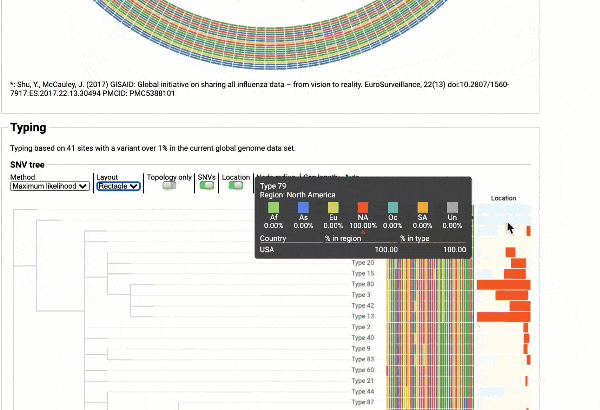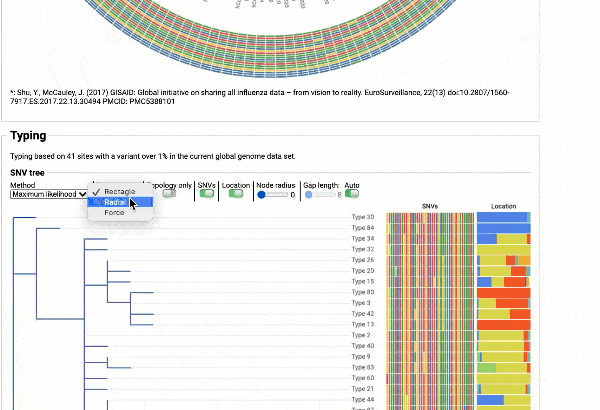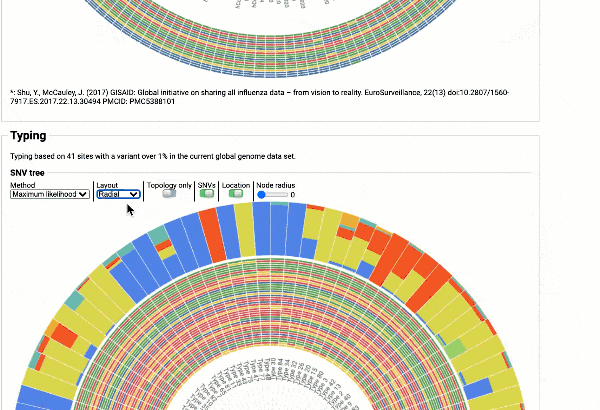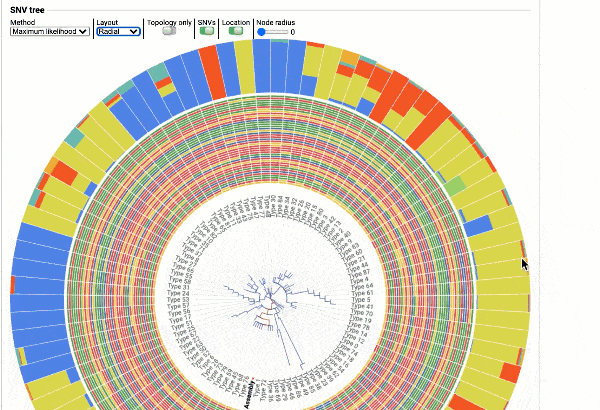
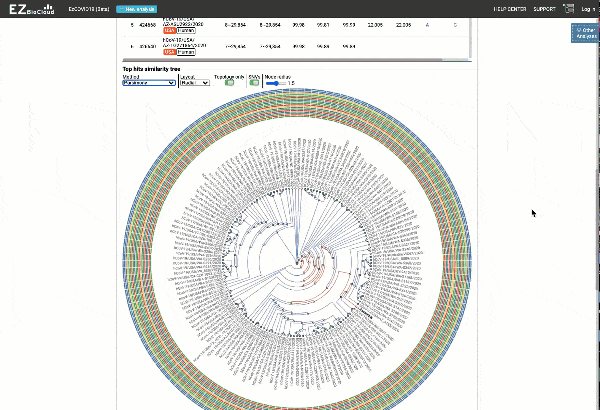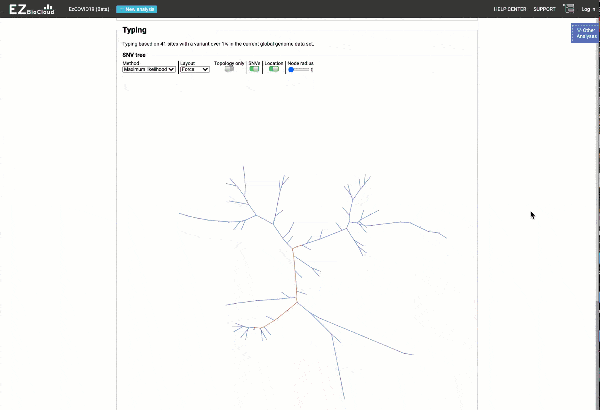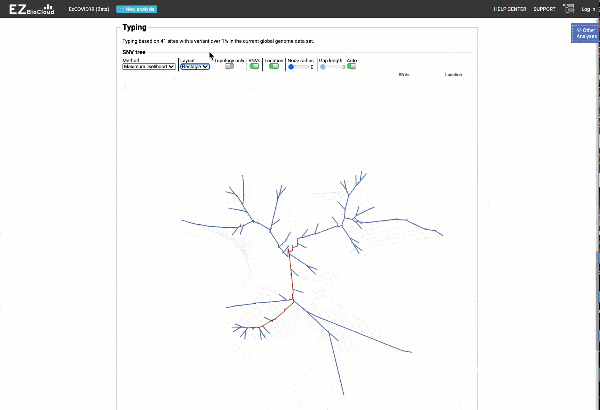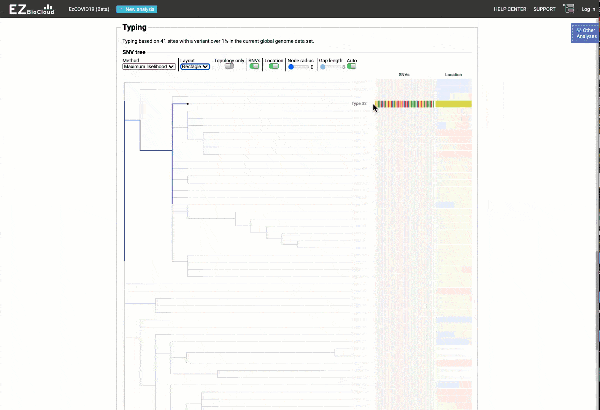
EzCOVID19 identifies the most similar genomes available in the reference databases (i.e., GISAID)
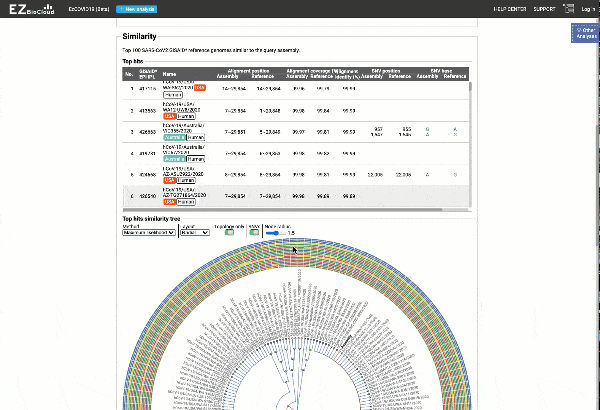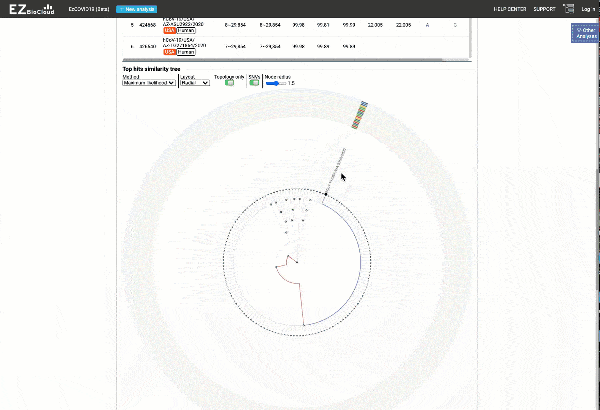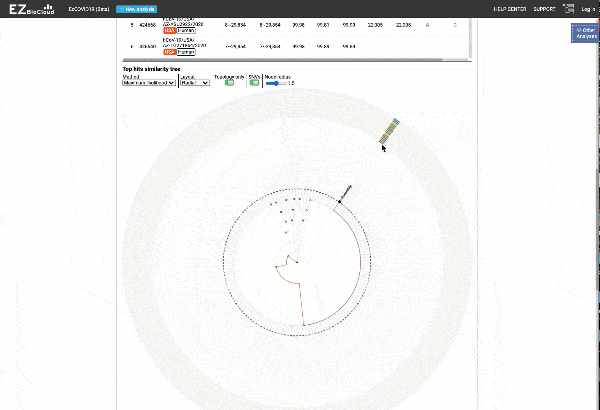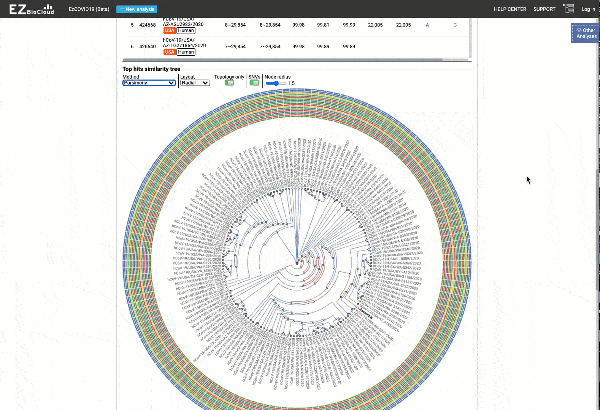
Including a maximum likelihood or parsimony based similarity tree
Obtain classification or typing of the queried genome using EzBioCloud’s SNP based classification scheme of SARS-CoV-2 variants
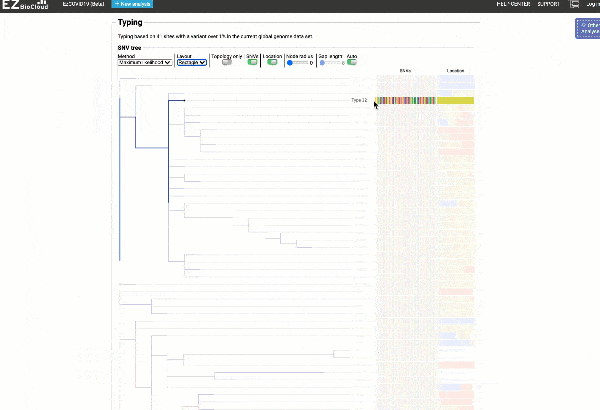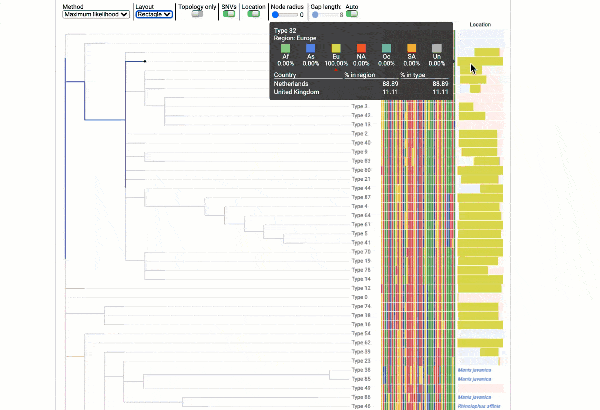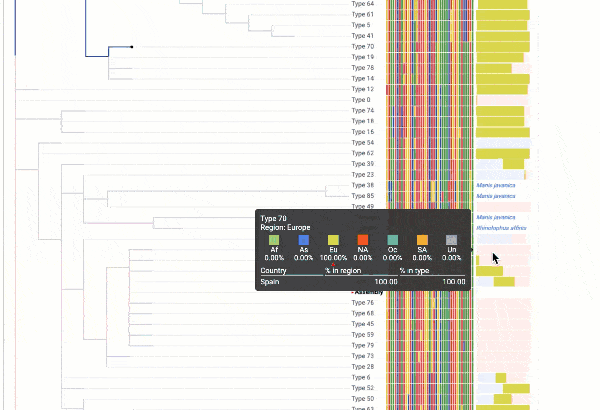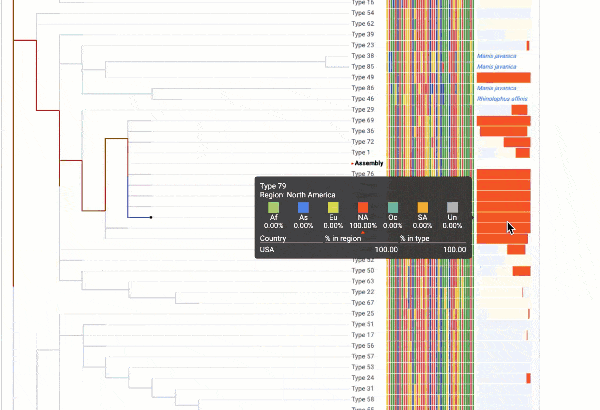
Including an evolutionary analysis of the detected SARS-CoV-2 type along with other types observed among publicly available SARS-CoV-2 genomes.