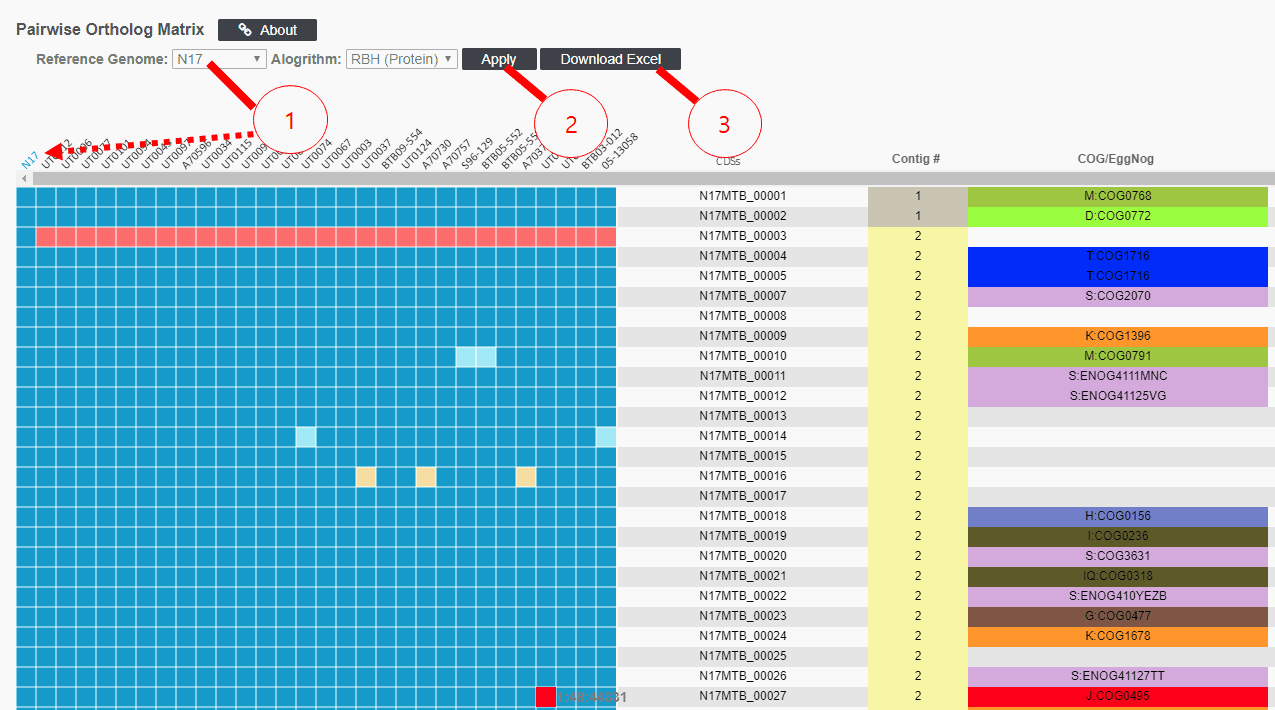
The pairwise ortholog matrix (POM) table was generated by a complete calculation of pairwise ortholog detection in a given dataset. If there are 10 genomes in a set, 10 by 10 calculations are carried out, excluding calculations for self-comparisons. For ortholog detection, an algorithm called Reciprocal Best Hit (RBH) is used to generate the table. The scientific background of ortholog detection is given here. The details of POM table are explained below.
| 1 | Select the “Reference genome” here. Orthologs are displayed in the direction of Reference → Subject genome. In this case, the Reference genome is displayed in the first column ( the whole column has 100% bit score ratio as a result of self-comparison). |
| 2 | Click [Apply] to save your choice of a reference. The contents of the table will remain unchanged until this button is clicked. |
| 3 | Click [Download excel] to save the whole table in excel format. |
Columns in the POM table
| CDS name: | Displays the name of CDS in the reference genome. |
| Contig #: | Index of contigs of the reference genome. An index is a unique number that starts from 1. Note that contigs #1 and #2 do not necessarily mean they are adjacent. |
| Location: | Location of CDS on the contig. |
| Gene: | Gene names and other synonyms for CDS. |
| Product: | Inferred name of translation product (protein). Usually, this information is theoretically derived and not experimentally confirmed. |
| The remaining columns: | Strain names of each genome |
Updated on May 15th 2016 (EK)