Species-level microbiome analysis using EzBioCloud
EzBioCloud 16S database is designed to be best performed for species-level identification even though there is a limitation due to the lack of sequence differences in some closely related species. The combination of EzBioCloud 16S DB and sensitive bioinformatics pipelines allows us a species-level exploration of human microbiome data as well as environmental samples.
Human microbiome project data
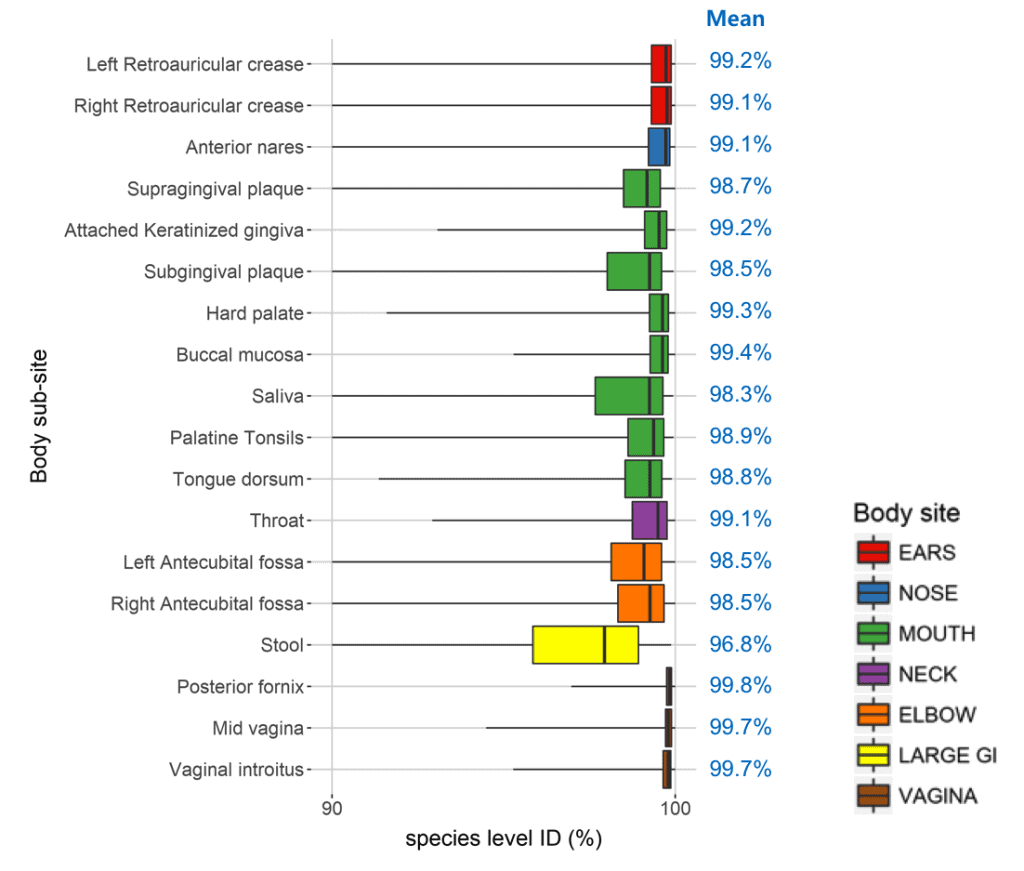
We performed the EzBioCloud MTP pipeline with EzBioCloud database on the Human Microbiome Project data (8,048 samples) that hold bacterial composition of 19 body sites of healthy people. The taxonomic coverage of EzBioCloud DB at the species level ranged from 96.8% (stool) to 99.8% (vagina). If you want to check individual samples, please follow this tutorial here.
A Faecalibacterium case
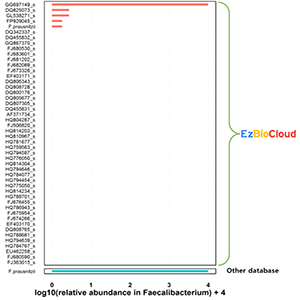
Let me bring up a case the genus Faecalibacterium for a more specific example. This genus is known as a major player in the human gut environment. Unfortunately, there is only one species that is officially recognized, that is, Faecalibacterium prausnitzii. In EzBioCloud, this genus is further classified into 48 species on the basis of 16S and genome sequences [Learn more]. Below are the comparisons of bacterial composition between EzBioCloud and one of the other databases.
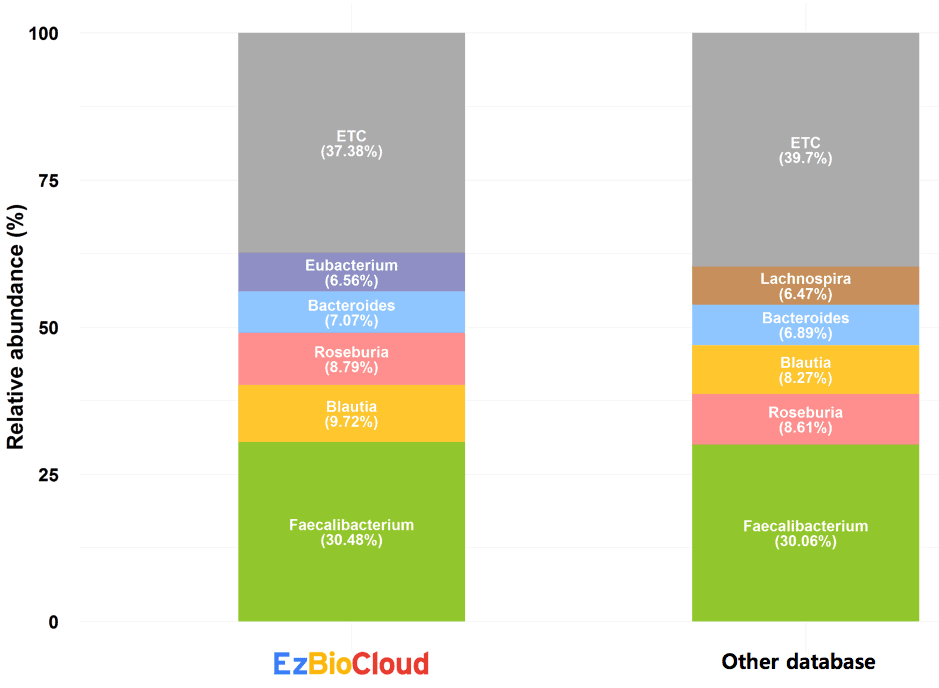
Below is the compositional difference of a fecal microbiome sample between EzBioCloud and other databases. The genus level compositions are very similar for Faecalibacterium.
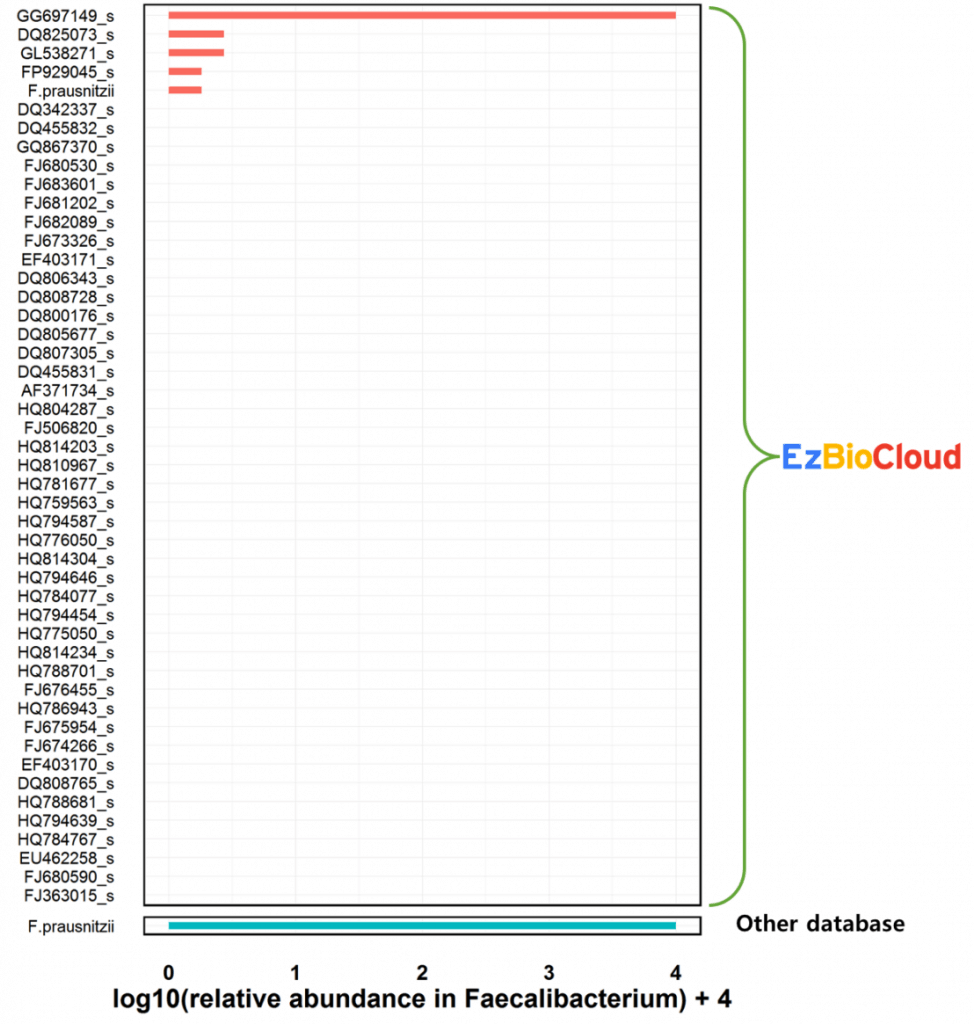
However, species-level compositional differences are dramatic (as below chart). Using EzBioCloud, we can identify 5 different Faecalibacterium species. Genome sequence data are available for some species [Learn more].
A Bacteroides case
Another human fecal sample rich in the genus Bacteroides, also a major inhabitant of the human gut. Again, compositions at the genus level are similar.
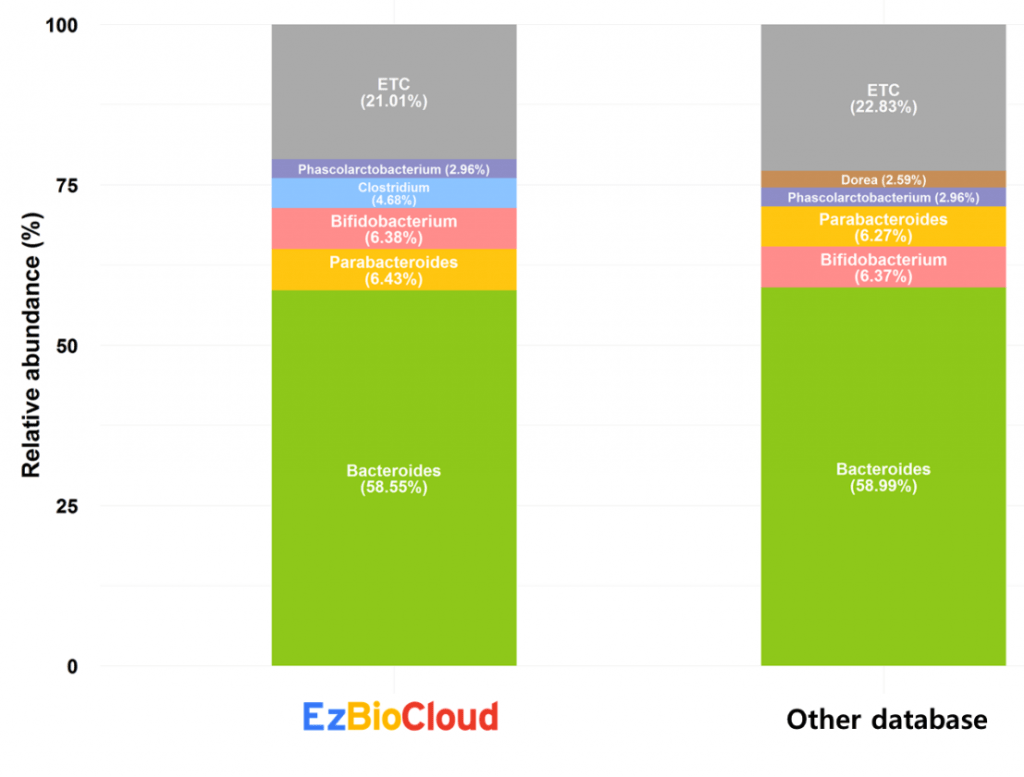
However, species-level compositions are significantly different from results between two databases. Please note that EzBioCloud has species which are recently described as well as uncultured phylotypes such as DQ798855_s.
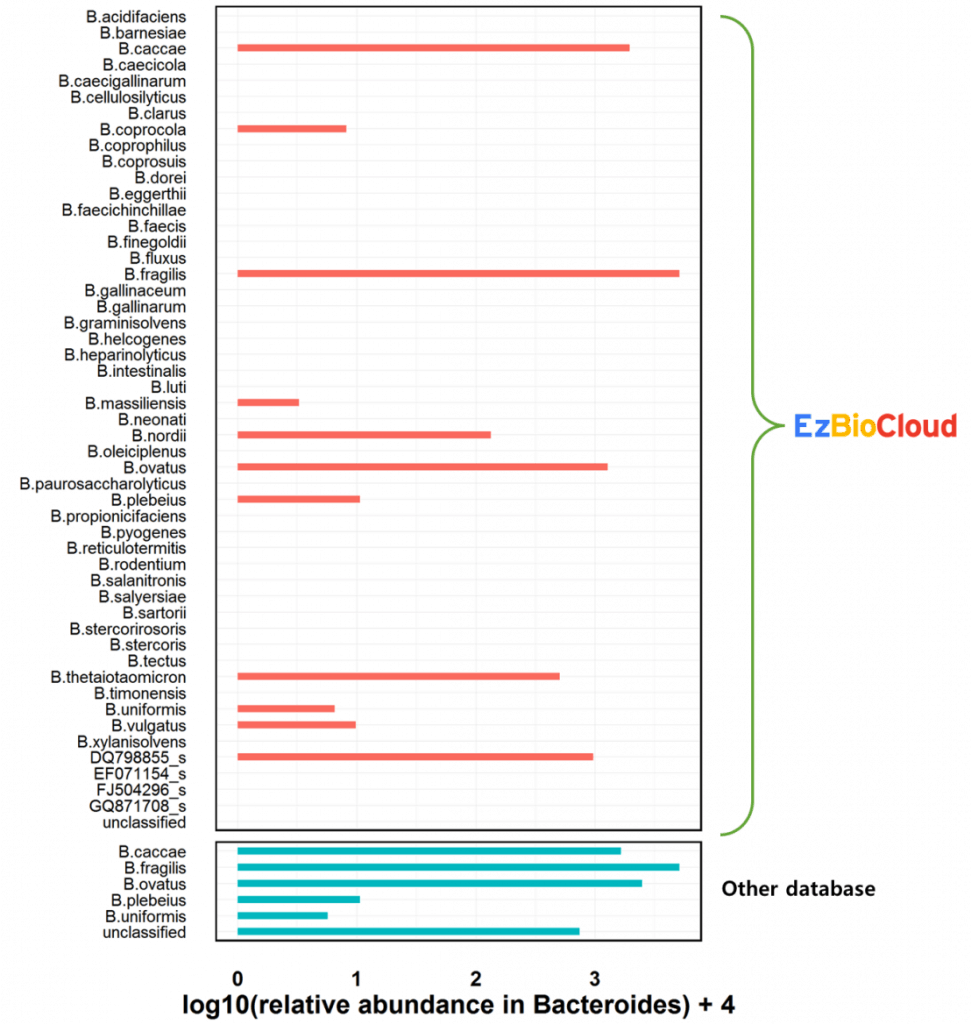
The current EzBioCloud 16S database, ver. mtp1.5, contains 272 species of the genus Bacteroides and 557 species of the genus Prevotella. It is far larger than any other database at the species level classification.
To obtain your copy of EzBioCloud 16S database for QIIME/MOTHUR pipelines, please visit here.
Last edited on Aug 10, 2017 (JC)