Technical document from CJ Bioscience, Inc.
Title: Experimental protocol for 16S rRNA gene-based microbiome sequencing using Illumina iSeq 100 Sequencing System
Document version: 1.0
Background
The Illumina iSeq 100 Sequencing System is a benchtop NGS instrument that is suitable for 16S rRNA gene-based microbiome analysis. For more details information about the instrument, please consult the Illumina’s website.
Sequencing accuracy:
The sequencing accuracy is an important factor for amplicon sequencing of mixtures such as microbiome. The iSeq 100 System uses new sequencing chemistry, called One-Channel SBS Chemistry, that is different from the Illumina MiSeq System. According to Illumina and our own experience, the iSeq 100 System produces higher quality reads than the Illumina MiSeq and other higher throughput instruments [Learn more].
Read length:
The iSeq 100 System can be used for paired-end sequencing (150 bp x 2). Unlike the MiSeq System, we can also sequence 16S amplicons using single-end (300 bp x 1) sequencing protocol which is described in this document.
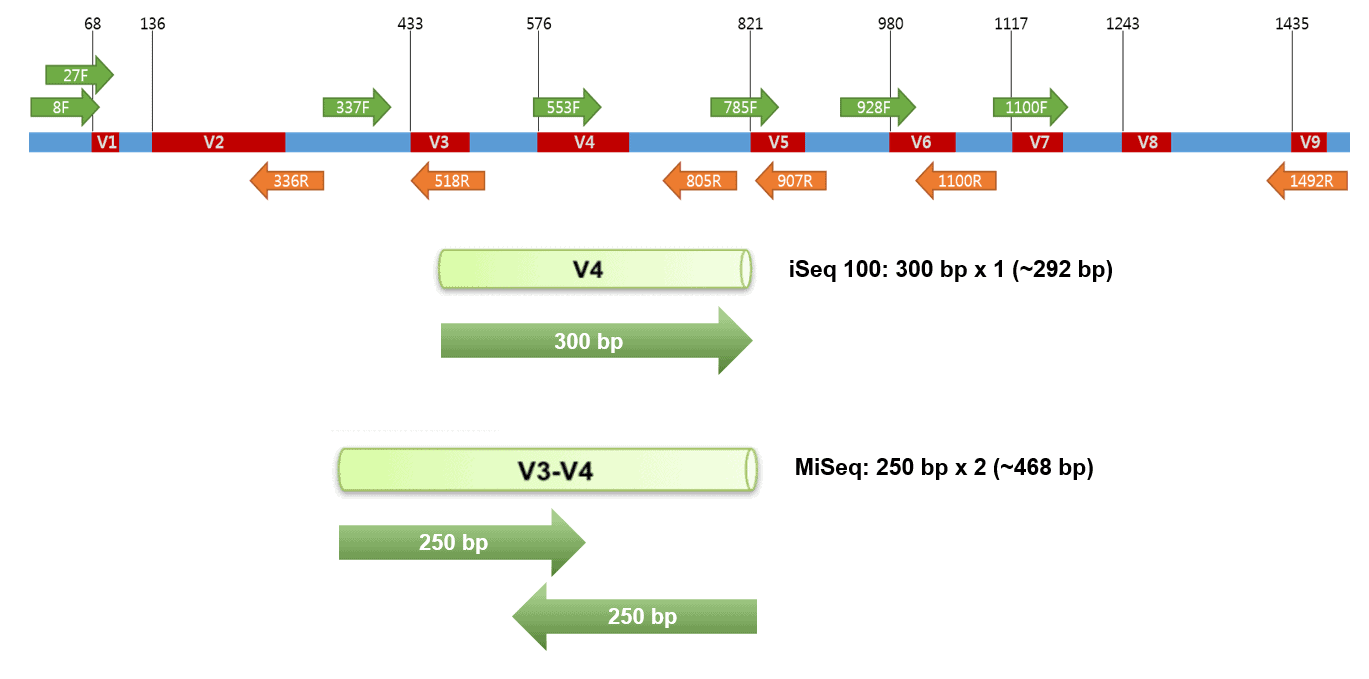
Sequencing region:
The 16S rRNA gene is about 1,500 bp long. The gene contains several conserved regions that are suitable as PCR priming sites. Therefore, various regions have been targeted for different NGS instruments depending on read-lengths and chemistry [Learn more]. We recommend the V4 region for the iSeq 100-300 bp x 1 sequencing protocol. This region has been most widely used, which can maximize comparability with the publicly available dataset. The length of the V4 region of E. coli K12 is 292 bp including the amplification primers.
Protocol
DNA preparation:
- Metagenomic DNA samples are prepared using the preferred methods or kits.
- Quantify a DNA concentration using Quat-iT PicoGreen dsDNA Assay kit (Invitrogen), follow the manufacturer’s instruction.
PCR primers:
These PCR primers target the V4 region of bacterial 16S rRNA gene.
Fusion primers are constructed according to the following combinations.
- 515F fusion primer (3’-P5 + i5 + NexTera consensus + Sequencing adaptor + Target region-5’)
- 806R fusion primer (3’-P7+ i7 + NexTera consensus + Sequencing adaptor + Target region-5’)
Download the sequence information of the multiplexed fusion primer set that includes the barcodes.
PCR amplification:
Setting up the PCR reaction
9 hours, not exceeding 18 hours
| Reagents | Volume (µl) |
| Taq Buffer | 2.5 |
| 2.5mM dNTP Mix | 2.0 |
| 515F Primer (10 pM) | 1.0 |
| 806R Primer (10 pM) | 1.0 |
| Taq Polymerase (5U/ µl) | 0.25 |
| PCR-grade Water | 16.25 |
| Template DNA | 2.0 |
| Total reaction volume: | 25.0 |
Thermocycler condition
Typical PCR reaction is given below. You may modify for your own needs.
| Step | Temp. (C) | Time | |
| Initial denaturation | 95 | 3 min | |
| Denaturation | 95 | 30 sec | 25 cycles |
| Annealing | 55 | 30 sec | |
| Extension | 72 | 30 sec | |
| FInal extension | 72 | 5 min |
Cleaning up and pooling
- Run amplicon from samples on gel electrophoresis or similar instruments (such as LapChip system). Expected size for PCR product is approximately 500 bp.
- Clean-up and size selection to each PCR product using AMpure bead XP (Beckman Coulter).
- Quantify an each purified PCR product with Quat-iT PicoGreen dsDNA Assay kit.
- Combine an equal amount of amplicon from each sample (50-100 ng) into a single tube.
- Clean-up and size selection to pooled amplicon using AMpure bead XP (Beckman Coulter).
- Measure final concentration with Quat-iT PicoGreen dsDNA Assay kit and A260/A280 ratio. For best result, the ratio should be between 1.8–2.0
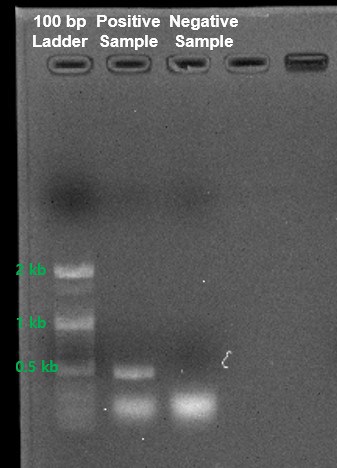
(1% Agarose gel, 1X TAE Buffer, 100V 25 min)
Sequencing library construction
- Measure library (pooled sample) size by Agilent 2100 Bioanalyzer (Agilent Technology) using Agilent DNA 7500 kit.
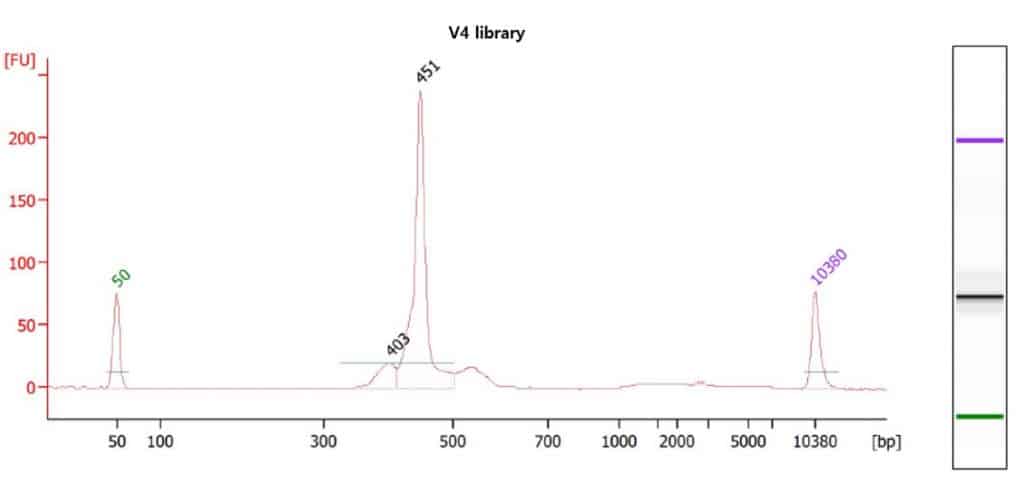
(DNA7500 Kit)
- For optimizing cluster densities, quantify the libraries using a fluorometric quantification method that uses dsDNA binding dyes or qPCR.
- For qPCR, KAPA Library Quantification Kit and KAPA standard kit (Kapa Biosystems) are used.
Sequencing
- Remove the iSeq100 cartridge from -25°C to -15°C storage and thaw the bagged cartridge using one of the following methods. After thawing, do not refreeze.
| Method | Thaw Time |
| 20°C to 25°C water bath | 6 hours, not exceeding 18 hours |
| 2°C to 8°C refrigerator | 36 hours, not exceeding 72 hours |
| Room-temperature air (20°C to 25°C) |
9 hours, not exceeding 18 hours |
- Dilute concentrated final library using Resuspension Buffer (RSB) to 1nM, and dilute to final loading concentration (Amplicon: 50 pM, Genome: 200 pM)
| Library Type |
Loading Concentration (pM) |
1 nM Library Volume (µl ) |
RSB Volume (µl ) |
| AmpliSeq for Illumina Library Plus | 50 | 5 | 95 |
| Nextera DNA Flex | 200 | 20 | 80 |
| PhiX (for a PhiX-only run) | 60 | 6 | 94 |
- Combine the following volumes in a E.P tube to dilute PhiX to the same loading concentration as the library. Add 2 to 10% of diluted PhiX to diluted sample library (Final volume 20 ul)
|
PhiX Loading Concentration |
1 nM PhiX Volume |
RSB Volume |
| 50 | 5 | 95 |
| 200 | 20 | 80 |
- Using a new pipette tip, pierce the Library reservoir and push the foil to the edges to enlarge the hole.
- Add 20 μl diluted library to the bottom of the reservoir.

- Open the white foil package from the notches. Insert the flow cell into the slot on the front of the cartridge.
- Avoid touching the electrical interface, CMOS sensor, glass, and gaskets on either side of the glass.

- In the control software, select Sequence from the Home screen.
- The software opens the door at an angle, ejects the tray, and initiates the series of run setup screens.
- Place the cartridge onto the tray so that the access window faces up and the flow cell is inside the instrument.
- Select Close Door to retract the cartridge and close the door.

Examples of iSeq 100 data
ATCC Microbiome Standards MSA-1002
- To get the detailed information about this Standard, visit ATCC’s website.
- We generated two iSeq 100 sequencing files with the protocol given in this document. To download them, click [File1] [File2].
- You can upload these files to EzBioCloud.net following the instruction given here.
- Sequencing data of ATCC Microbiome Standards from Illumina MiSeq (V3V4), iSeq 100 (V4) and PacBio (V1V9=full length) are pre-loaded at www.ezbiocloud.net. The below video will explain how to browse them and incorporate with your own data.
Human fecal sample
- We generated two iSeq 100 sequencing files with the protocol given in this document. To download them, click [File1] or [File2].
- The following link will let you explore the bacterial composition of a human fecal sample. https://www.ezbiocloud.net/iseq100_fecal
Last updated Feb. 12, 2019



