A short survival guide for microbiologists
How are names controlled?
- Bacterial nomenclature and naming are regulated by the International Code of Nomenclature of Prokaryotes (shortly the Prokaryotic Code), but the classification of actual species is not. Please download the latest Code here. For example, the name Escherichia coli is regulated by the Prokaryotic Code, but not the properties and taxonomic classification of the species itself. This means that there is no official taxonomy and only names are controlled. Therefore, the terms ‘valid species’, ‘formal species’ or ‘official species’ do not make sense. Only the name can be validated so ‘species with a validly published name’ is OK to use. I indicated this in red as there are so many mistakes in the literature.
- As for animals and plants, bacterial (and archaeal) nomenclature follows the binomial nomenclature introduced by Carl Linnaeus.
How is a species name formed?
- A species name should have two parts as a binomial system: (1) generic name and (2) specific epithet. This combination represents the ‘species name’ and it should be unique in the nomenclature system.
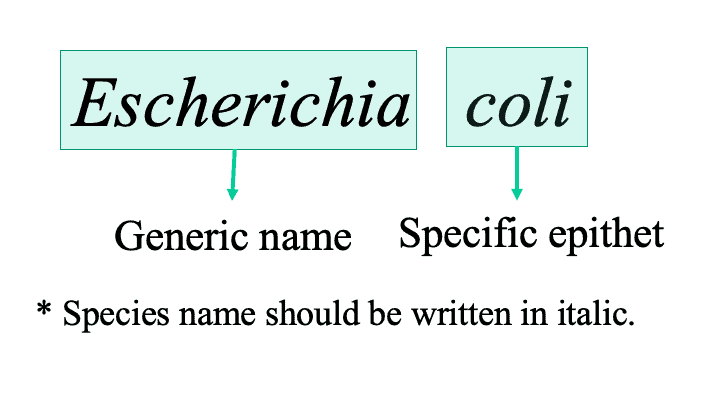
- The scientific names of all taxa must be treated as Latin; names of taxa above the rank of species are single words.
Names are organized into a hierarchical system
- The hierarchical system of the official nomenclature is as follows: (only the popular ones are given)
- Phylum (or Division): at present, the phylum rank is not controlled by the Prokaryotic Code. Oren et al. proposed to include ‘phylum’ in the Code, but the proposal was not yet discussed and approved by the International Committee on Systematics of Prokaryotes (ICSP), the body that controls the Prokaryotic Code [Learn more].
- Class: The type of a class is one of the orders. The class is named after the type genus of the type order of the class.
- Order: The type of an order is one of the genera. The order name is named after the type genus of the order. E.g. The order Pseudomonadales is named after the type genus Pseudomonas.
- Family: In general, the family name is named after the type genus of the family, e.g. The family Pseudomonadaceae is named after the type genus Pseudomonas.
- Genus
- Species
- Subspecies: Subspecies are created only when it is necessary. There is no ‘type subspecies’ concept in the Code. I think that many subspecies should be re-evaluated using genomics as they are mainly classified by single or a few phenotypes. For example, an important probiotic species, Lactobacillus delbrueckii contains six subspecies.
| Rank | Suffix | Example |
| Order | -ales | Pseudomonadales |
| Suborder | -ineae | Pseudomonadineae |
| Family | -aceae | Pseudomonadaceae |
| Genus | Pseudomonas | |
| Species | Pseudomonas aeruginosa |
- These names are given by the scientists who propose the species and assign it to a genus. Naturally, any species can be assigned in this hierarchical system; however, this is not always implemented. Officially, a species must be classified in a known genus but assigning that genus to a family, then an order and so on is optional. Frequently this happens because the taxonomic relationships are ambiguous, so it isn’t really clear how to group genera into higher taxonomic ranks. This leaves a lot of chaos in the bacterial taxonomic system and it’s particularly bad news for microbiome researchers where investigations are made at the different taxonomic ranks such as family or even phylum.
- For example, the genus Thermicanus has been proposed without its assignment to a known family (Gössner et al. 1999). In NCBI taxonomy, it is under a tentative family ‘Bacillales Family X. Incertae Sedis‘ under the order Bacillales. EzBioCloud database assigns every species under the complete hierarchical system, so Thermicanus was assigned under a tentatively created family ‘Thermicanus_f‘ (please note that it is an invalid name and is not italicized). Of course, anyone can propose the family Thermicanaceae by publishing the proposal.
How can a name be validated (more precisely, validly published)?
- As mentioned previously, the names are controlled by the Prokaryotic Code. To get a name validated, the name should be included in one of the lists described below.
- Approved List of Bacterial Names:
- This was published in 1980 by Skerman, McGowan, and Sneath, signified a new start in prokaryotic nomenclature.
- From this List, any subsequent changes are published by International Journal of Systematic and Evolutionary Microbiology (IJSEM) (formerly International Journal of Systematic Bacteriology (IJSB)).
- There are two types of updated Lists published in IJSEM, namely ‘Notification List’ and ‘Validation List’.
- Notification List:
- IJSEM is the official journal of the International Committee on Systematics of Prokaryotes (ICSP) and the Bacteriology and Applied Microbiology Division of the International Union of Microbiological Societies (IUMS). This means that IJSEM is the official journal for prokaryotic nomenclature. Please also note that there is no official bacterial taxonomy.
- Any taxonomic proposals that include changes to names (new taxa or changes of names) published in IJSEM will automatically be reviewed by the IJSEM’s List Editors. If the paper meets the requirements of the Prokaryotic Code, the proposed changes will be listed in ‘Notification List’ and are officially ‘validated’.
- Please see this example.
- Validation List:
- The Prokaryotic Code also allows scientists to validate names effectively published in journals other than IJSEM.
- Once a name is published, it is called ‘effectively published‘.
- Anyone who cares about the science should make sure that any effectively published names be validated by sending the publication pdf to IJSEM with proof that (for species and subspecies) the designated type strain is available without restrictions from at least two public culture collections located in different countries. This can be done by anyone including authors. Consequences of failing to do this can be damaging, as the name is ‘effectively published’ but never ‘validated‘. The List Editors of the IJSEM will add the names in a Validation List on condition that all requirements of the Prokaryotic Code are met (for new species and subspecies notably Rules 27 and 30).
- This is an example of the Validation List.
- Approved List of Bacterial Names:
- Invalid names: Unfortunately, there are several cases where a name could not be validated.
- A name was used widely before 1980, but not included in the Approved Lists of Bacterial Names in 1980
- A name was published outside of IJSEM, but never validated as either:
- No one sent the pdf and further required documentation to IJSEM for validation.
- The publication does not meet the minimal requirements of the Code. These minimal requirements will be explained later.
- Names not validly published are given in quotation marks, e.g. ‘Selenomonas massiliensis’ or “Selenomonas massiliensis” to differentiate from validly published names.
- The minimal and only requirements for validation of names of new species and subspecies according to the Prokaryotic Code are:
- A type strain must be designated. Sometimes, a paper describing a new species does not mention which strain is the type strain. In such cases, the name cannot be validated.
- The type strain should be deposited to two culture collections in two different countries. This ensures that the type strain remains available also for example when a culture collection discontinues its activity or loses the strain.
- The type strain must be available to anyone through the culture collections. Patent strains cannot serve as type strains. If you want to patent a strain and restrict the distribution, it cannot serve as the nomenclatural type for a species or subspecies.
- A proper etymology and description should be given (see below).
Why using the right names is important
- Bacterial names are designed to be scientifically organized to give biological insights based on phylogeny; this is possible using the hierarchical taxonomic system. For example, we think that the species Escherichia coli shares more phenotypic characters with other Escherichia species than with a Bacillus species. Therefore, using the right names is critical in interpreting microbiome and other types of data.
- Unfortunately, there are still many names that are in the wrong places in the current nomenclature. For example, Clostridium scindens has been known for its important role in bile acid metabolism of human gut microbiota and still has the name of Clostridium. However, it is not even close to the genus Clostridium and should be classified in a new genus in the family Lachnospiraceae. The authentic Clostridium would be placed under the family Clostridiaceae.
- A few other examples related to the human microbiome are given below from our past blog posts:
Candidatus
- In some cases, the basic requirements of the Prokaryotic Code cannot be satisfied even though a species is real and scientifically important. Under the formal nomenclature, these species will never get their name validly published. So, taxonomists created the term ‘Candidatus‘ to support the tentative naming.
- For Candidatus names, there are no pure cultures, or isolates, therefore no type strains.
- Candidatus names are not governed by the Prokaryotic Code, so they do not belong in the formal nomenclature thus are not ruled by the Prokaryotic Code.
- A typical example is Candidatus Pelagibacter ubique
- This species contains uncultured or cultured bacteria from the ocean and is probably the most abundant species on earth, so it must be very important [Learn more].
- The reason that it has never been validated is that the species can be cultured, but not in a way, or scale that can be used by culture collections for long-term storage. Hence, the minimal requirement of the Code cannot be fulfilled. In addition, the specific epithet (ubique) is malformed.
- Not all Candidatus taxa are well characterized as the rank of Candidatus is not regulated by the Prokaryotic Code.
- I think that the minimal standards for Candidatus are:
- Availability of full-length high-quality 16S sequence as this gene is the framework of the taxonomy of Bacteria and Archaea; this is a must!.
- Availability of genome sequence that is not contaminated and reasonably complete (checked by the presence of domain-level core genes).
- Minimal ecological and metabolical traits should be provided.
- Anything else can be subjectively decided. Nonetheless, Candidatus is a tentative name at the time of writing.
How to name a new taxon
- It is best if the name can represent the typical characters of the species. For example, the name Plasticicumulans means “accumulating plastic”. The genus was thus named because the type species accumulates the bioplastic polyhydroxyalkanoate.
- Another popular way of naming a new taxon is to name after a person. This person should have a record of contributing to science. It is unethical to name after yourself, to a member of your family or to a person who is not related to science (e.g. the President of your country). The famous Escherichia and Salmonella were named after the microbiologists Escherich and Salmon.
- Naming after geographical location is also popular, but not encouraged (even though I did it many times).
- You will need an etymology for your new taxon. I suggest consulting a specialist.
- Read this article by the late Hans G. Trüper for the further details.
Tips for writing a taxonomic paper describing new species
- Anyone who is working on isolation of bacteria should have a good chance of finding a new species. Using genome sequencing, I would say that your chance of finding new species from one isolation study from soil or water should be almost 100%. It is our duty, as good microbiologists, to make them known to the scientific world by describing them as new species.
- Assuming that you have isolated multiple strains, designate the type strain of your new species. If you have a single strain, that strain will become the type strain of the new species.
- Prove it is a new species by 16S rRNA gene (16S) and genome sequencing
- Start with 16S sequencing and obtain a full-length, high-quality sequence. Don’t trust the assembled sequence from sequencing firms, as these service companies almost never edit the final sequence. Always check the chromatograms (Sanger ab1 files) manually. I have seen terrible 16S sequences used for creating a new species, which are not really new species.
- Search the sequence against the EzBioCloud 16S database at www.ezbiocloud.net. If you find any hit lower than 98.7%, you probably have a new species given that your sequence is of good quality [Learn more].
- Let’s say you have ~99% 16S similarity, don’t be disappointed. You still have a very good chance of finding a new species. I suggest you go for whole genome sequencing. For the taxonomic purposes, a guideline was published here. Calculate Average Nucleotide Identity (ANI) values between your isolate and type strains of closely related species in the 16S searches using EzBioCloud database. The ANI cutoff is 95~96% for species.
- If you have indeed confirmed that you have a new species, try to obtain diagnostic phenotypic data. This can vary depending on the taxa.
- Obtain the general morphological and physiological characteristics
- Obtain the differential characteristics that can be used for separating your new species from others. This data is usually given as a Table (e.g. see Table 1 of this paper as an example).
- Bergey’s Manual is a great resource to obtain phenotypic data of related species [Learn more].
- Please add the whole genome sequence of type strain in your study.
- Like the 16S gene, the genome sequence is becoming the taxonomic basis for all disciplines of microbiology. If you do not sequence your new genome, you are leaving a hole in the system which must be filled by somebody else.
- Currently, Syst. Appl. Microbiol. and IJSEM require the genome sequence of the type strain at least for any new classification.
- Deposit the type strains to two culture collections in two different countries.
- To find suitable culture collections, use the WFCC site.
- My recommendation is to find culture collections that at least have an English website.
- Make sure that you get the certificate of the deposition from two culture collections in two countries. If they don’t know what the certificate of the deposition is, don’t bother and try to deposit to other culture collections. This process takes several months, so start as soon as you find that you have a new species.
- Write the taxonomic proposal as a manuscript.
- As with your usual content in any paper, a proper etymology and description should be given (see below as an example).
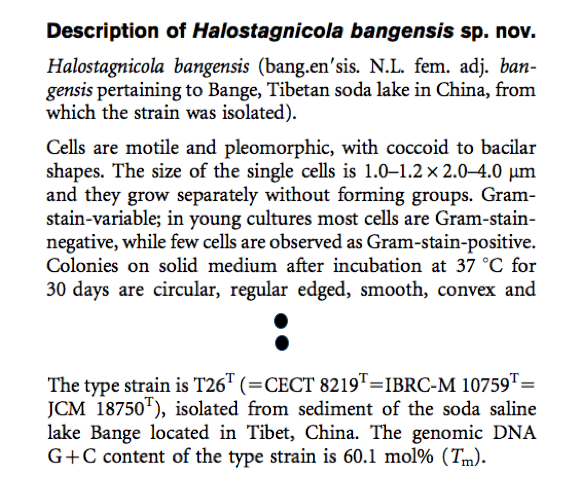
- Submit the manuscript to IJSEM with two certificates of deposition of the type strain. It is OK to submit to other journals as long as they publish the papers in English. If it is published outside of IJSEM, please make sure that you submit the pdf of the publication plus the two certificates of deposition to IJSEM for validation. Without validation, your new species will not be properly acknowledged by other scientists. In fact, in many cases, it adds more confusion than good.
Who controls all of this process?
- Basically, international committees of microbiologists are maintaining the Code (as the official rulebook) and the official journal (IJSEM) which publishes the Notification Lists and the Validation Lists.
- The most important committee is the International Committee on Systematics of Prokaryotes (ICSP). ICSP is the body that oversees the nomenclature of prokaryotes, determines the rules by which prokaryotes are named, and whose Judicial Commission issues Opinions concerning taxonomic matters and revisions to the Prokaryotic Code, etc.
- The members and Executive Board members of the ICSP are elected from the representatives of each country’s microbiological societies. If you are interested in participating in any process of prokaryotic nomenclature and taxonomy, you need to be elected as a representative from a microbiological society and attend these events (ICSP meetings). Please visit ICSP’s website for more details. I believe that ICSP would love to have more young microbiologists engaged in the process of bacterial taxonomy (remember that you are required to be a representative of your society first).
Resources for up-to-date nomenclature
- Please note that only the Approved Lists of Bacterial Names of 1980, and the Notification and Validation Lists in the IJSB and IJSEM represent the official nomenclature. However, IJSEM or other officially recognized bodies do not provide an organized, up-to-date list of validly published names at present. Fortunately, the following resources are maintained properly and are therefore useful. Please bear in mind that these websites may contain human errors.
Quiz for students
Please use NCBI, EzBioCloud or related taxonomic and sequence databases.
- Firmicutes is an extensively studied phylum in the human microbiome. Most of the people have over 10% in their gut. Is it controlled by the official nomenclature of bacteria (the Code)? If so, why?
- Write a brief report about Candidatus Liberibacter asiaticus with the following information:
- Taxonomic history
- Biology of this species
- Genomes and ecology
- What is the Latin meaning of specific epithet, salmonicida? How many bacterial species have the same specific epithet of salmonicida? Is it correct if more than two species have the same specific epithet? Find two more species names with salmonicida as a species epithet?
- Nocardia pinensis is a bacterial species that causes the serious problems in activated sludge in Australia. The name of the species was later changed. Find out this nomenclatural change and why this was necessary (on what scientific basis).
I appreciate the comments from Barny Whitman, Ramon Rosselló-Móra and an anonymous colleague.
This document is a part of my course – Prokaryotic Taxonomy & Microbiome.
by Jon Jongsik Chun, CEO of CJ Bioscience Inc.
Last edited on Feb. 3, 2019