In this tutorial, we will work you through how to use EzEditor2 for the alignment of protein-coding genes.
We will use an example file named “Leuconostoc_gyrB_unaligned.ezb” which contains gyrase subunit B gene sequences of the genus Leuconostoc. Please download this file from here.
Codon-based alignment
A gene encoding a protein has an open reading frame(ORF) that can be translated into an amino acid(AA) sequence. Therefore, we can obtain either nucleotide(NT) or amino acid sequences from the same DNA sequence. Since an AA sequence is the final product that is under evolutionary pressure, multiple alignment of protein-coding genes should utilize the AA sequence while aligning NT sequences. Codon-based alignment, or AA-aware alignment, is a method to align NT sequences via AA sequence alignment.
The general process is:
- Translate all NT sequences to AA sequences.
- Carry out a multiple alignment process with AA sequences.
- Insert gaps into NT sequences with the guidance of aligned AA sequences. Each gap in the AA sequence is equivalent to 3 gaps in NT sequences.
Multiple alignment using CLUSTAL-omega
EzEditor2 does not contain an algorithm for multiple alignment. Therefore you will need to install the CLUSTAL-omega program in advance. Please see here if you haven’t done this.
1. Open “Leuconostoc_gyrB_unaligned.ezb” file with EzEditor2, and click “Align Window” button.
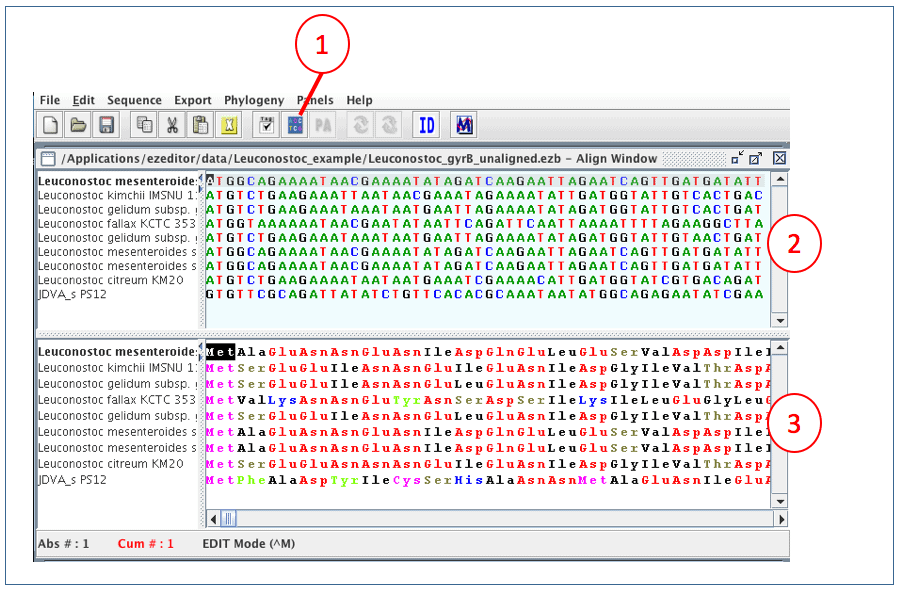
(1) “Align Window” button
(2) A panel showing NT sequences
(3) A panel showing translated AA sequences
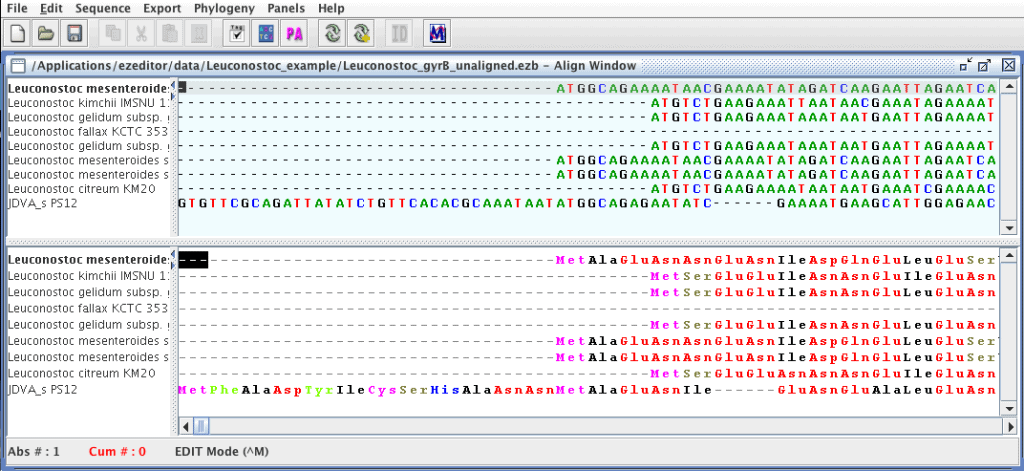
2. Execute codon-based alignment using the menu [Sequence -> Align by CLUSTAL-omega using codons]. After running this program, you should see the aligned sequences as below:
You may adjust the final alignment as you wish by inspecting manually.
Also, see:
Go to “Working with 16S rRNA sequences using EzEditor2”
Go to “Phylogenetic analysis using EzEditor2”
Go back to the content of tutorials
Last modified on January 12, 2017 (JC)